Back in May 2019, I wrote about a program called WGS Extract to produce from your Whole Genome Sequencing (WGS) test, a file with autosomal SNPs in 23andMe format that you can upload to sites like GEDmatch, Family Tree DNA, MyHeritage DNA, or Living DNA.
The mastermind behind this program, who prefers to remain anonymous, last month made a new version available.You can get it here: https://wgsextract.github.io/ The program last year was 2 GB. This one now is 4.5 GB. The download took about 45 minutes. And that is a compressed zip file which took about 3 minutes to unzip into 8,984 files totaling 4.9 GB. It didn’t expand much because the majority of the space was used by 5 already compressed human genome reference files, each about 850 MB:
- hg38.fa.gz
- hs37d5.fa.gz
- GCA_000001405.15_GRCh38_no_alt_analysis_set.fna.gz
- human_g1k_v37.fasta.gz
- hg19.fa.gz
I don’t know the technical aspects about what’s different in each of these references, except that 1 and 3 are Build 38 and 2, 4 and 5 are Build 37. For genealogical purposes, our DNA testing companies use Build 37.
Also included among the files and very useful are raw file templates from various companies which include the majority of the SNPs from each of the tests:
- 23andMe_V3.txt (959286 SNPs)
- 23andMe_V4.txt (601885 SNPs)
- 23andMe_V4_1.txt (596806 SNPs)
- 23andMe_V5.txt (638466 SNPs)
- 23andMe_V5_1.txt (634165 SNPs)
- MyHeritage_V1.csv (720922 SNPs)
- MyHeritage_V2.csv (610128 SNPs)
- FTDNA_V1_Affy.csv (548011 SNPs)
- FTDNA_V2.csv (720449 SNPs)
- FTDNA_V3.csv (630074 SNPs)
- FTDNA_V3_1.csv (613624 SNPs)
- Ancestry_V1.txt (701478 SNPs)
- Ancestry_V1_1.txt (682549 SNPs)
- Ancestry_V2.txt (668942 SNPs)
- Ancestry_V2_1.txt (637639 SNPs)
- LDNA_V1.txt (618640 SNPs)
- LDNA_V2.txt (698655 SNPs)
There are 4 summary files:
- 23andMe_SNPs_API.txt (1498050 SNPs) which likely combines the SNPs from all five 23andMe tests.
- All_SNPs_combined_RECOMMENDED_hg19_ref.tab.gz (2081060)
- All_SNPs_combined_RECOMMENDED_GRCh37_ref.tab.gz (2081060)
- All_SNPs_combined_RECOMMENDED_hg38_ref.tab.gz (2080323)
The last 3 appear to be a combination of all the SNPs from all the raw file templates. The hg19 and GRCh37 files appear to be the same, but differ in how the chromosome is specified, as 1 or as chr1, as MT or as chrM. I’m not sure how the hg38 file was derived, but it may have been a translation of all addresses from Build 37 to Build 38, excluding 737 SNPs that are in Build 37 but not Build 38.
Running WGS Extract
The program can now run on any computer. Just run the appropriate script:
- Linus_START.sh
- MacOS_START.sh
- WIndows_START.bat
Since I have Windows, I ran the third script. I had to tell Microsoft Defender SmartScreen to allow it to run. It starts up a Command window which then starts up the WGS Extract Window:
There are now three tabs: “Settings”, “Extract Data” and “Other”. Above is the Settings Page.
Here is the Extract Data page:
The Mitochondrial DNA and Y-DNA functions are both new.
And this is the Other page:
All the functionality on this 3rd page is new.
Two WGS Tests
When I first checked it out WGS Extract last year, I only had my Dante Short Reads WGS test. See: Creating a Raw Data File from a WGS BAM file. Since then, I have taken a Dante Long Reads WGS test.
Three BAM Files
The raw reads from a WGS test are provided as FASTQ files. These need to be put into the correct place on my genome. A file containing the mappings of each of my reads to where it is in my genome is called a BAM file (Binary Sequence Alignment Map). It’s these BAM files that WGS Extract reads.
I have 3 BAM files I can use:
- The BAM file Dante provided with my Short Reads WGS test. They used a program called BWA (the Burrows-Wheeler Aligner) to produce my BAM.
- Dante did not provide a BAM file with my Long Reads WGS test. So I did the alignment myself using BWA to produce a BAM from this test. I documented that in my Aligning My Genome post.
- I found out that the program minimap2 produced more accurate alignment than BWA for Long Reads. I tried to run that myself but the job was taking too long. Then I heard that YSeq offered the mapping service using minimap2, so I had them create a minimap2-based BAM file from my Long Reads WGS test.
Let’s now try a few things.
Show statistics
From on the Settings page, we first load our BAM file and select an output directory. Loading the huge BAM file is surprisingly quick, taking only about 5 seconds.
We can now go to the “other” page and press “Show statistics on coverage, read length etc.”
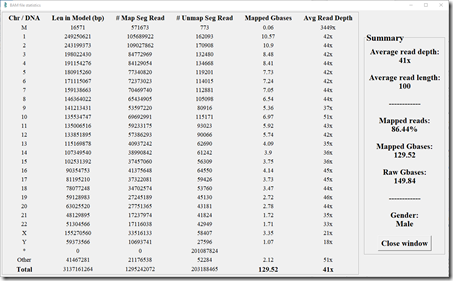
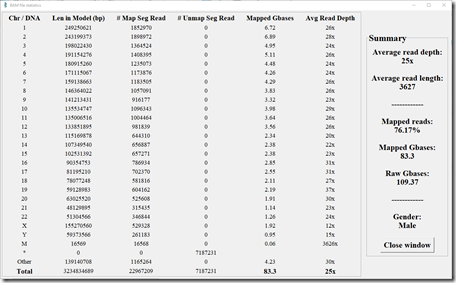
Here’s my statistics from my Short Reads test. (Click image to enlarge)
My Short Reads test consisted of almost 1.5 billion reads. 86.44% of them were able to be mapped to a Build 37 human reference genome. That gave an average of 41x coverage over every base pair. The average read length was 100 base pairs.
Here’s my statistics from my Long Reads test with BWA mapping:
Long Reads WGS test are known to have a higher percentage of errors in them than a Short Reads WGS test. But because their reads are longer, they still can be mapped fairly well to the human genome.
I had over 20 million long reads. 76.17% of the reads were able to be mapped, which is lower than the 86% from my short read test. This resulted in an average coverage of 25x versus the 41x from my short read test. The average read length of the mapped reads was 3627 base pairs, which is 36 times longer than my short read test.
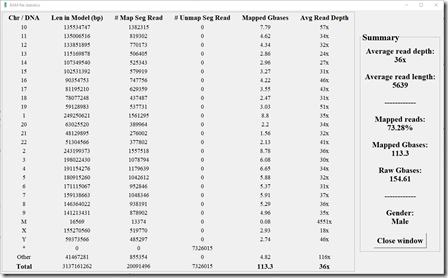
Here’s the stats from my Long Reads test aligned by YSEQ using minimap2:
I have no idea why the Samtools stats routine decided to show the chromosome in alphabetical order just for this run but not the other two above. That is definitely strange. But the stats themselves seem okay. This does show the improvement that minimap2 made over BWA since the average read depth is now up to 36x and the average read length of mapped reads is increased to 5639. I expect that BWA must have had trouble aligning the longer reads due to the errors in them, whereas minimap2 knows better how to handle these.
Haplogroups and Microbiome
First I run Y-DNA from the “Other” page using my Long Reads. WGS Extract now includes a version of the python program Yleaf which is available on GitHub to perform this analysis. It takes about 5 minutes and then gives me this.
That’s interesting. I know I’m R1a, but my more detailed groups from the various companies then start taking me into M198, Y2630, BY24978 and other such designations. I’ve not seen it strung out as a R1a1a1b2a2b1a3 designnation before. At any rate, it doesn’t matter too much. My Y-DNA does not help me much for genealogy purposes.
For Mitochondrial DNA, WGS Extract gave me this: 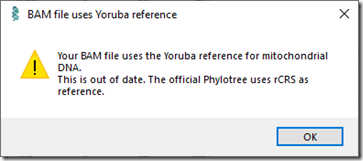
That’s okay. I already know my mt haplogroup is K1a1b1a. It doesn’t help me much for genealogical purposes either.
There was also an option in WGS Extract to create an oral microbiome that can be uploaded to app.cosmosid.com. This option will extract your unmapped reads which might be bacterial. I’m not interested in this so I didn’t try it.
Creating A DNA Raw Data File
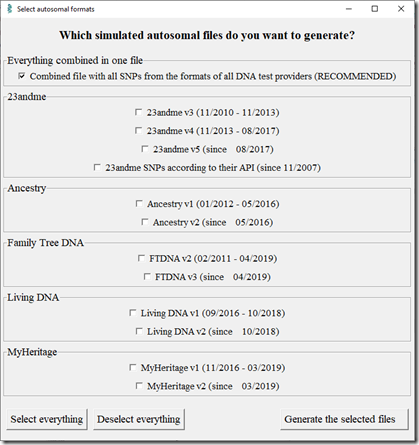
Going to the Extract Data page in WGS Extract, now I press the “Generate files in several autosomal formats”. It gives me this screen:
When the screen first popped up, everything was checked. I clicked “Deselect everything” and then checked just the combined file at the top.
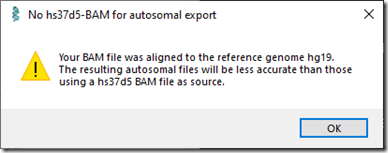
I did this for my Short Reads BAM file, When I pressed the Generate button at the bottom, the following info box popped up.
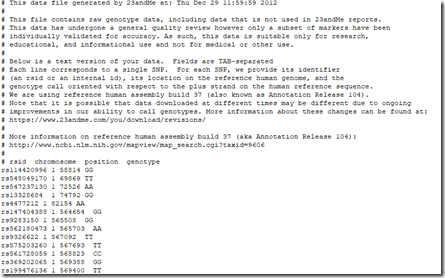
I pressed OK and the run started. After 65 minutes it completed and produced a text file with a 23andMe raw data header and over 2 million data lines that looks like this:
It also produced a zipped version of the same file, since some of the upload sites request compressed raw data files.
A Glitch and a Fix
I wanted to do the same with my two Long Read BAM files. When I tried, it was taking considerably longer than an hour. So I let it run all night. It was still running the next morning. It was still running in the afternoon.Why would a Long Reads BAM file take over 20 times longer than a Short Reads BAM file to run? They both are about the same size. The Long Reads file of course has longer reads but fewer of them.
I started wondering what was going on. I contacted the author. I posted about this on Facebook and got some helpful ideas. Finally I found the temporary files area that WGS Extract used. I was able to tell that for my Long Read BAMs, the temporary file with the results was not being created. I isolated the problem to the program mpileup that was the one failing. I searched the web for “mpileup long reads nanopore” and found this post: mpileup problem with processing nanopore alignment. It suggested to use the mpileup –B option.
The mpileup –B option stands for “no-BAQ”. The Samtools mpileup documentation explains BAQ to be Base Alignment Quality. This is a calculation of the probability that a read base is misaligned. Allowing the BAQ calculation “greatly helps to reduce false SNPs caused by misalignments”.
I tried adding the –B option, and now WGS Extract worked! It took 75 minutes to run for my BWA Long Reads file and 115 minutes for my YSEQ minimap2 Long Reads file. I then ran my Short Reads file with the –B option and it ran in only 20 minutes. I’ll compare that last run with my Short Reads run with the –B option, and that should give me an estimate as to how many false SNPs might have been introduced.
Next Steps
I’ll compare these 4 WGS Extract files with each other and with my 5 raw data files from my standard DNA tests in my next blog post. I’ll see if I can determine error rates, and I’ll see how much I can improve the combined raw data file that I’ll upload to GEDmatch.