I was wondering in my last post if I could create a raw data file that could be uploaded to to GEDmatch or DNA testing company from my Whole Genome Sequencing (WGS) results. I was trying to use one of the Variant Call Format (VCF) files. Those only include where you vary from the human reference. So logically you would think that all the locations not listed must be human reference values. But that was giving less than adequate results.
Right while I was exploring that, there was a beta announced for a WGS Extract program. It works in Windows and you can get it here:
This is not a program for the fainthearted. The download is over 2 GB because it includes the reference genome in hg19 (Build 37) and hg38 (Build 38) formats. It also includes a windows version of samtools which it runs in the background as well as the full python language.
I was so overwhelmed by what it brought that I had to ask the author how to run the program. I was embarrassed to find out that all I had to do was run the “start.bat” file that was in the main directory of the download, which opens up a command window that automatically starts the program for you, bringing up the screen I show above.
WGS Extract has a few interesting functions, but let me talk here about that one labeled “Autosomes and X chromosome” with the button: “Generate file in 23andmeV3 format”. I selected my BAM (Binary Sequence Alignment Map) file, a 110 GB file I received by mail on a 500 GB hard drive (with some other files) from Dante. I pressed the Generate file button, and presto, 1 hour and 4 minutes later, a raw data file in 23andMe v3 format was generated as well as a zipped (compressed) version of the same file.
This was perfect for me. I had already tested at 5 companies, and had downloads of FTDNA, MyHeritage, Ancestry, Living DNA and 23andMe v5 raw data files. I had previously combined these 5 files into what I call my All 5 file.
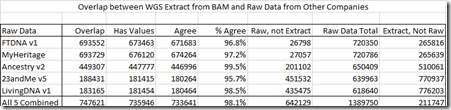
The file WGS Extract produced had 959,368 SNPs in it. That’s a higher number of SNPs than most chips produce, and since it was based on the 23andMe v3 chip, I knew there should be quite a few SNPs in it that hadn’t been tested by my other 5 companies.
You know me. I did some analysis:
The overlap (i.e. SNPs in common) varied from a high of 693,729 with my MyHeritage test, to a low of 183,165 with Living DNA. These are excellent overlap numbers – a bit of everything.
Each test had a number of no-calls, so I compared all the other values with what WGS Extract gave me, and there was a 98.1% agreement. That’s a 2% error that is either in the chip test, or in the WGS test, but from this, I cannot tell whether its the chips or the WGS that are the incorrect values. But in each case, one of them is.
When I compare this file to my All 5 file, which has 1,389,750 SNPs in it, I see that there are an extra 211,747 SNPs in my WGS file. That means I’ll be able to create a new file, an All 6 file, that will have 1,601,497 SNPs in it.
More SNPs don’t mean more matches. In fact they usually mean fewer matches, but better matches. The matches that are more likely to be false are the ones that get excluded.
In addition to including the new matches, I also wanted to update the 747,621 SNPs in the file to the same SNPs in my All 5 file. As noted in the above table, I had 2,305 SNPs whose values disagreed, so I changed them to no calls. No calls are the same as an unknown value, and for matching purpose, are always considered to be a match. Having more no calls will make you more “matchy” and like having less overlap, you’ll have more false matches. The new SNPs added included another 905 no calls. But then, of the 20,329 no calls I had in my All 5 file, the WGS test had values for 9,993 of them.
So my number of no calls went from:
20,329 + 2,305 + 905 - 9,993 = 13,546, a reduction of 6,783.
I started with 20,329 no calls in 1,389,750 SNPs (1.5%),
and reduced that to 13,546 no calls in 1,601,497 SNPs (0.8%)
A few days ago, I was wondering how much work it take to get raw data for the SNPs needed for genealogical purposes out of my WGS test. A few days later, with this great program, it turns out to be no work at all. (It probably was a lot of work for the author, though.)
I have uploaded both the 23andMe v3 file, as well as my new All 6 file to GEDmatch to see how both do at matching. I’ve marked both research. But I expect once the matching process is completed, I’ll make my All 6 file my main file and relegate my All 5 file back to research mode.
Here are the stats at GEDmatch for those who know what these are:
WGS Extract SNPs: original 959,368; usable 888234; slimmed 617,355
All 5 SNPs: original 1,389,750; usable 1,128,146; slimmed 813,196
All 6 SNPs: original 1,601,497; usable 1,332,260; slimmed 951,871