I received the rest of my raw data files for my WGS (Whole Genome Sequencing) test today. It was shipped from their lab in Italy and came on a 1 TB hard drive.
Previously I was able to download my VCF files from Dante’s site. I reported on the files a couple of months ago in my post: My Whole Genome Sequencing. The VCF File. Those files were compressed and totaled 224 MB and expanded to 869 MB. The VCF (Variant Call Format) files only contain the variants, i.e. the readings where I vary from the human genome reference.
The files supplied this time include all my data, not just the variants. So they are much larger. As a result they were sent to me on the large hard drive. They include the BAM and FASTQ files.
The files are provided in three folders:
- clean_data
- result_alignment
- result_variation
The FASTQ files
The clean_data directory contains 16 files named something like:
aaaaaaaaaaa_L01_5mm_n.fq.gz
where aaa is some identifier, and mm run from 78 to 85 and n is 1 or 2.
Each file is about 8 GB in size and is gzip compressed at about 34%. I have a fast Intel i7 computer and it takes 30 minutes to uncompress one of these files to its full size of about 22 GB.
When unzipped, the .gz drops off the file name and the .fq suffix represents a FASTQ file.
Here’s what the beginning of one of the unzipped FASTQ files looks like:
Shown above are the first 4 groups of readings in one of the files, where 4 lines make up a reading. The first line of each group of 4 is an identifier, the 2nd are exactly 100 base pari values, the 3rd is just a plus (at least in the records I glanced at) and the 4th are codes that represent the quality of each of the 100 reads.
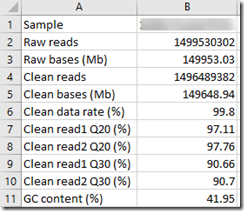
Also in the directory is a small excel file that contains some summary statistics from my WGS test. It contains:
Sample is my kit number. There were 1.5 billion reads. In order to get an average of 30x coverage on 3 billion base pairs, I figure the average read length would have to be 60 base pairs.
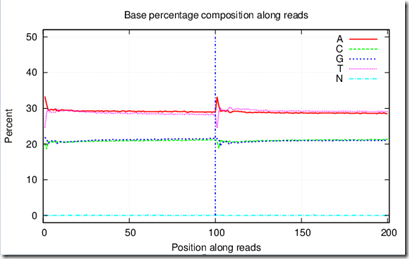
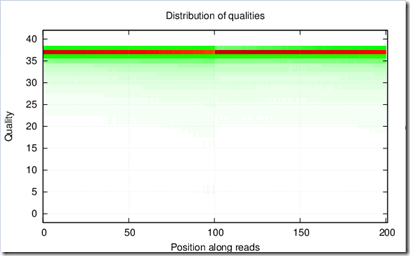
Each of the eight 5mm files have two pictures associated with them that show some results, e.g.
aaaaaaaaaaa_LO1_578.base.png contains:
and aaaaaaaaaaa_L01_578.qual.png contains:
Most of this is all new to me too, so I can’t explain what all this means yet.
The BAM file
The result_alignment directory contains the aaaaaaaaaaaaaa.bam file. It is 115 GB in size compressed at 27% which expands to 425 GB. Decompression time for this file on my computer is over 17 hours. For most purposes, you don’t want to decompress this file since most genome analysis programs work with the BAM file itself and with a small bam.bai (BAM index) file of 8 MB that is also in the directory. The BAM index file allows the programs to go directly to the section of the genome that the analysis program needs.
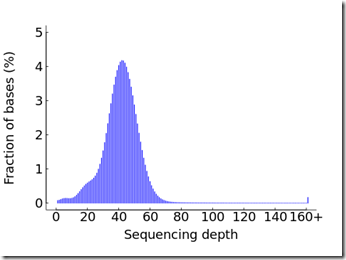
This directory also contains the same summary xls file that was with the FASTQ files (see above) and three png images:
aaaaaaaaaaa.Depth.png that shows that I mostly achieved at least 30x coverage
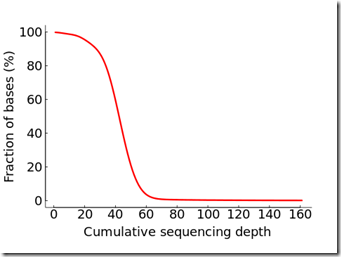
aaaaaaaaaaa.Cumulative.png that shows the cumulative distribution of the depth
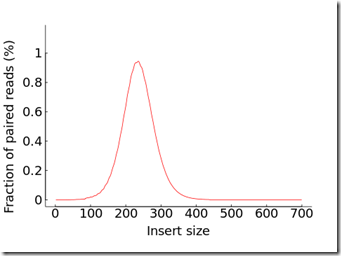
aaaaaaaaaaa.Insert.png
Insert size has something to do with the analysis process. I used to know what paired reads are, but I forgot. I’ll have to look that up again if I ever have to use them.
Variation Files
There are four subdirectories named sv, snp, indel and cnv:
They contain a number of files, some gzipped (which I decompressed in the above listing). Basically, these are various files indicating all my gene, exome and genome differences from the human reference for both my SNPs and my INDELs (insertions/deletions). These files are in a different format than the two VCF files I downloaded for SNPs and INDELs earlier.
What’s Ahead
I purchased the Long Read WGS test from Dante a few days ago. I think I’m going to wait until I get the results from my Long Read test. This will likely take a few months. Once I get the long read results, I’ll look at the BAM and FASTQ files from both tests, compare them to each other, and see what I can learn from them.
With just one test, you can’t tell how good it is. But with two, you can compare their results to each other. It should be interesting.



Joined:
1 blog comment, 0 forum posts
Posted: Tue, 23 Apr 2019
Hello, Did you pay an extra fee to receive the raw files on hard drive? or was that cost included in the cost of ordering the WGS? Since I am disabled with a few rare disorders, I am very interested in WGS for medical reasons, not genealogy. but I appreciate your sharing your explorations of the WGS and reviews of Dante Labs’ tests. I look forward to hearing more about the Long Reads WGS. I already have Dante’s 30x WGS, and I am wondering about any additional insights that the Long Reads test might yield for my very complex conditions.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Tue, 23 Apr 2019
Kendraz: When I purchased my kit back in August 2018, the hard drive with the BAM and FASTQ files was included free of charge. I did note in November that they started charging $69 extra for the hard drive containing your raw data.
I am purely looking at the short versus long reads for the purpose of determining the accuracy of the reads, determining ways to correct the SNPs, as well as the unlikely possibility to phase the pair of chromosomes into the two parents. My interest is aiding genetic genealogists. I have little interest in or knowledge about the medical side of it.