There’s a bad rumor going around that segment matches, especially for small segments, can be very old. I’ve heard expectations that the segment might come from a common ancestor 20 generations back or even 30, 40 or more. And that’s said to happen even if you have a fairly large 15 cM segment.
Part of this is due to the incorrect thinking that a segment of your DNA has been around forever and has been passed down from some ancient ancient ancestor to you and to just about everyone else. Since there is only a 1/2 chance that each generation gets the segment from the right parent, the argument is that it gets offset maybe by the more than 2 children per generation that keep the segment alive all the way down to two 30th or 40th generation descendants who then happen to share the segment. That also assumes there is no intervening ancestor along some other path who is more recent than that 30th generation one. For endogamy, the argument is that the segment has proliferated through the people and most of them happen to have it. Although in that case, I find it hard to believe that there is not a line to a different common ancestor who is fewer than 30 generations back.
The fallacy here is that all our DNA segments are ancient. They are not. In fact, many of them are quite recent, only a few generations old.
Let’s take a look at, say a 15 cM segment that you got from your father. You could have:
1. Got the whole segment from your father’s father’s chromosome,
2. Got the whole segment from your father’s mother’s chromosome, or
3. There could have been a recombination that occurred somewhere along the 15 cM segment and you got part of it from your father’s father and part from your father’s mother.
It is case number 3 that is interesting. In this case, that 15 cM segment is no longer the same as your father’s father’s segment, nor is it the same as your father’s mother’s segment. It is a new segment that has been born in you and you are the first ancestor to have that segment and maybe you’ll pass it down to many of your descendants. And no one else will have that segment that you have, unless some random miracle as rare as a lottery winning happens.
Also, your father’s father’s segment at this location and your father’s mother’s segment both are not passed down to you. Maybe they’ll be passed to a sibling of yours or maybe they won’t. But both of your grandparent’s segments have died along your line.
So what actually happens is that any segment of your DNA has its birth in one of your ancestors. That ancestor may pass it down to zero or more descendants, and if it is passed down, each descendant may or may not continue to pass it down. The segment eventually dies. A recombination on the segment can’t be avoided forever.
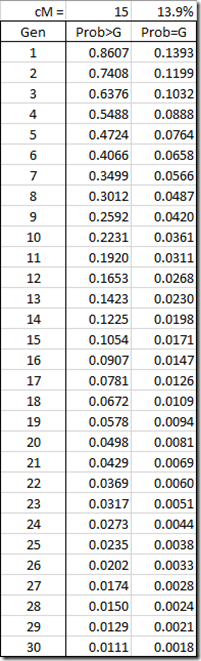
Now what is the probability of a new 15 cM segment being “born” in you? Well, that’s what cM represents and there will be about a 15% chance that any particular 15 cM segment of your DNA was formed from a recombination in your parent, and that you have a brand new segment. For most purposes, using the cM as a percentage is close enough. But for more accuracy, I’ll use the actual probability from the equation P(recomb) = 1 – exp(-cM/100) which gives 13.9%. (See my Update Jan 26, 2020 about this equation)
Well guess what? The probability that any particular 15 cM segment is born in any of your ancestors is also 13.9%. The chance that the segment was not born, but was passed down is therefore 86%. We can use that fact to now calculate the probability that this segment was passed down any number of generations to some descendant:
What this says is that if you have a 15 cM segment, then there is about a 50% chance that it was created in one of the last 5 generations, a 75% chance that it was created in one of the last 9 generations, and 95% chance that it was created in one of the last 20 generations. The average age of segments that size is 7.2 generations (1 / 13.9%). This is very simple mathematics/statistics.
If you match with another person on the same segment, then they have the same probabilities. The chance both of you got this segment from more than 20 generations back would be only 5% x 5% = 0.25%.
Revisiting Speed and Balding Once Again
I’m still frustrated that Speed and Balding’s simulation results are being used without question to estimate segment age for human DNA segment matches.
About two years ago, I used two different sets of calculations, one my own in Revisiting Speed and Balding, and one based on work by Bob Jenkins in Another Estimate of Speed and Balding Figure 2B. In both cases, I found segment age estimates that were somewhat less than Speed and Balding.
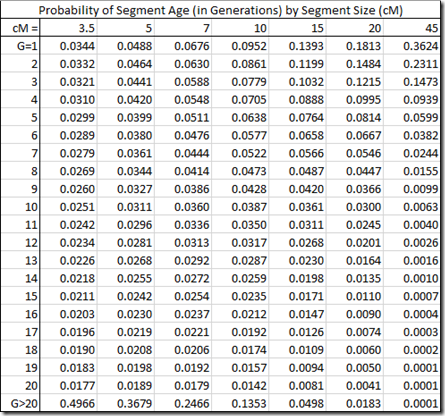
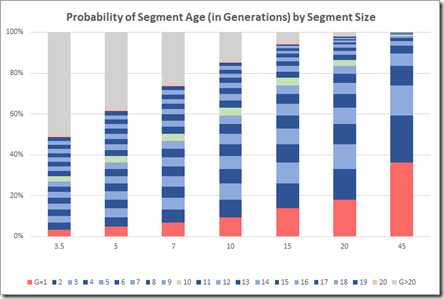
Let’s see how my Segment Life estimates compare. Picking a few different segment sizes and calculate their values gives:
And then lets plot these in a stacked chart:
Look at the gray area at the top left. That’s the probability of segments of the given segment size being 20 or more generations old. The green bar is the divider at 10 generations. You likely have a good chance to identify how you’re related to segment matches that are under the green bar, indicating that most segments over 15 cM should be identifiable and that even very small segments might be identifiable.
Compare this to Speed and Balding:
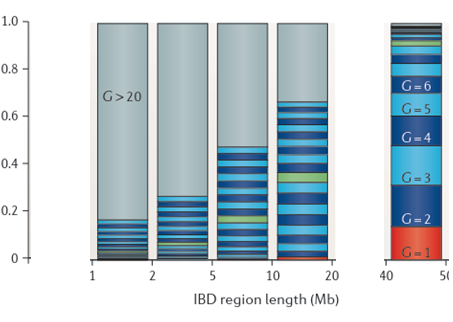
Speed and Balding give much larger chance of older segments than does my segment life methodology, or than do either of the two analyses in my earlier blog posts.
Conclusion
Segments aren’t passed down from ancient times. They are created and die all the time due to recombination events and they may not be as old as you are led to believe. Some of your smaller matching segments. e.g., between 5 and 15 cM have (by my segment life and other earlier calculations) a 40% to 70% chance of originating less than 10 generations ago. This means you might be able to determine how you’re related to your match.
By using triangulation techniques (such as Double Match Triangulator), you can determine triangulations of segments in the 5 to 15 cM range which will eliminate most by-chance matches. You can then put your segment matches into Triangulation Groups, to help find the common ancestor of the group and connect your DNA matches to your tree.
Update Jan 26, 2020: After discussion with Celia Baitinger on the Facebook Genetic Genealogy Tips and Techniques group, we realized that the Wikipedia equation for P(recomb) = (1 – exp(-2 * cM / 100) / 2 may only be for recombinations that involve an odd number of crossovers. For genetic genealogy, we are interested in all crossover events. As a result, the correct analysis should be this:
Assuming a Poisson distribution for crossovers (which is what is usually assumed), then the P(zero recombs) when the mean is cM/100 is: exp(-cM/100), and therefore:
P(recomb) = 1 - exp(-cM/100)
I have updated the figures in the above article to reflect this correction. No changes were significant enough to affect any of my observations or conclusion.

Joined: Mon, 21 Jan 2019
3 blog comments, 0 forum posts
Posted: Tue, 20 Aug 2019
Nice writeup, I’ve been waiting for someone to talk about this.
There’s a corollary that you didn’t talk about. Ancestry has a 6 cM minimum size for matching segments. Any segment under 6 cM might as well not exist, as far as Ancestry’s matching algorithms are concerned. Other sites use slightly different cutoffs, but I’ll stick with 6 cM for the moment.
That means that from Ancestry’s perspective, your example 15 cM segment _wasn’t_ born in some ancestor. It “got longer” relative to at least one 6+ cM segment from one of that ancestor’s parents. You can still trace through to the earlier, shorter segment from the earlier generation(s).
In contrast, an 11 cM segment might have been created from two 5.5 cM segments that _can’t_ be traced back to earlier generations when using a 6 cM minimum. A segment of that type really would seem to appear out of nowhere.
Can you extend your math to show the probability of one segment being traceable for X generations but NOT traceable further in its shorter form?
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Wed, 21 Aug 2019
Jonathan: I don’t understand your logic why a 6 cM limit means that the 15 cM wasn’t born in some ancestor, but got longer. It’s always a recombination that creates a new segment. Whether it or parts of it are filtered out by the company doesn’t seem relevant to me because the filtering is only done in the DNA tester’s results.
Joined: Mon, 21 Jan 2019
3 blog comments, 0 forum posts
Posted: Wed, 21 Aug 2019
I guess I’m hung up on “the 15 cM segment”. From what I’ve seen, it’s rare that three people share the EXACT same segment, with exact same start and end points. So when I see “the 15 cM segment”, I read “a 15 cM region of DNA, where various matches might share various overlapping bits within that region.”
Suppose that the 15 cM segment was “born” by combining two segments that were each exactly 7.5 cM long, and suppose that you match someone on the first 7.0 cM of the 15 cM segment. I can’t think of any way to tell from the DNA alone whether that 7.0 cM match was from a close relative where the 15 cM segment happened to get shorter, or from a distant relative that provided the original 7.5 cM segment that got extended to form the larger 15 cM segment.
Contrast that to a situation where you have a 10 cM segment that was born by combining two 5 cM segments. In that case you CANNOT have a match from an earlier ancestor, since a match to a 5 cM segment would be invisible to any matching algorithm that used a 6 cM minimum.
So I agree with you that the 15 cM segment WAS born in some ancestor. But I’m saying that if you can’t do anything with that information then it isn’t helpful.
The important parts are the recombination points. Any DNA match that shares a segment with you that crosses a recombination point must share with you the ancestor where that recombination happened. Of course, identifying each recombination point is much harder than simply talking about them! :-)
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Thu, 22 Aug 2019
Ahh. Now I see what you’re trying to do. You’re taking the two pieces of the 15 cM segment, and deciding that if one was only 5 cM, then it wouldn’t match anyone because the smaller side wouldn’t reach the threshold that the company calls their minimum match.Yes. That’s entirely possible and may be the reason why our more distant matches don’t always cover our entire genome.