Today Evert-Jan Blom, author of Genetic Affairs and the new clustering algorithm implemented by MyHeritage DNA, posted on the Genetic Genealogy Tips & Techniques group on Facebook. He announced some improvements to his AutoCluster analysis on Genetic Affairs for 23andMe matches.
He posted:
A well known feature for the DNA relatives list on 23andme are the Relatives in Common. What is interesting is that 23andme, just like MyHeritage, supplies the shared cM values between shared matches. On MyHeritage, we use this data to improve the analysis of people from endogamous populations.
In addition, there is a Shared DNA column in the Relatives in Common list. The Matches marked with a “Yes” have overlapping segments – and, according to the research of Jim Bartlett (https://segmentology.org/2017/…/20/triangulation-at-23andme/) over 99% of the time these matches form a Triangulated Group (TG).The shared cM values between matches as well as TG information is now employed for 23andme AutoCluster analyses. …
But what about these TG data? … The rectangles that contain a DNA helix symbol have overlapping segments and probably form a TG. I’ve already discovered some clusters that could be extended by taking into account some grey cells that in fact were TGs. …
So we supplement the ICW based 23andme AutoCluster analyses with TG data which already improves the analysis. And, although we know that not all members of a (large) cluster will form TGs, wouldn’t it be interesting to only take into account TG data? We thoughts so as well and therefore are this feature is now also available for 23andme AutoCluster.
I wrote about MyHeritage’s New AutoClustering feature 3 weeks ago, showing my results. Unfortunately, I don’t have any DNA matches at MyHeritage whose relationship I truly know, so I couldn’t identify the ancestral source of any of the clusters.
But at 23andMe, I have a number of known relatives who tested. So I should be able to identify some of the clusters. Let’s see how it goes.
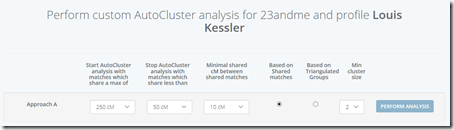
I went to my account at Genetic Affairs and added my 23andMe website to it. Then I requested an autocluster using the default parameters:
I performed an analysis first “Based on shared matches” and then did it again after selecting “Based on Triangulated Groups”
For the shared matches, I got all one big cluster of 54 members:
So that’s my endogamy and 1 cluster is not of much help. However, notice that some of the cells have a little DNA symbol in them, like this:
These are people who are not only ICW (In Common With) myself and the row person and column person, but also are shown in the 23andMe Relatives in Common list as a “Y” in the Shared DNA column. That mean’s that all three of us share at least one common segment of DNA with each other, i.e. we triangulate somewhere.
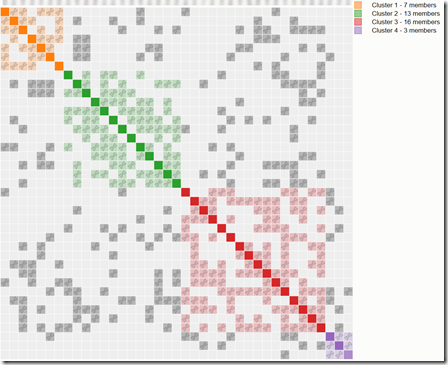
So Evert-Jan had the innovative idea to allow just the use of these triangulating people to be used for clustering. When my second run based on Triangulated Groups came back, it looked like this:
This initially got me really excited. There were just four clusters and I was hoping that this clustering had done the trick and divided my DNA relatives into my four grandparent groups. Did it?
The Trouble with Using Triangulations for Clustering
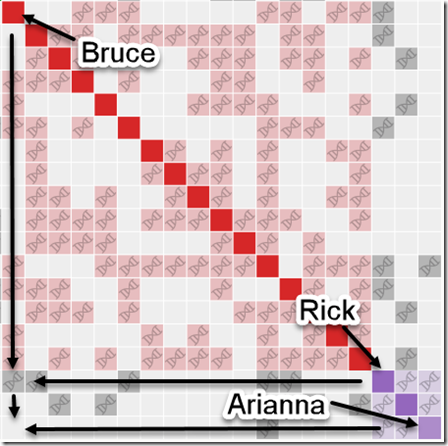
Unfortunately, I noticed something very important. The first person in the red group is Bruce, a 3rd cousin of mine. The first person in the purple group is Rick, his brother, also my 3rd cousin. If you go down the column of the first red box to the row of the first purple box, you can see the two of them have a grey square with DNA symbol in it meaning the three of us triangulate. We have two full brothers triangulate who absolutely must be in the same cluster no matter which way you look at it. So why aren’t they?
While I was looking at all of this, Evert-Jan himself Facebook messaged me, and we started discussing this problem. I then noticed that the 3rd purple box in the last row and last column with Rick in the purple group was Rick’s daughter Arianna. If you then look down from Bruce’s red box to Arianna’s row, you’ll see there is no triangulation!
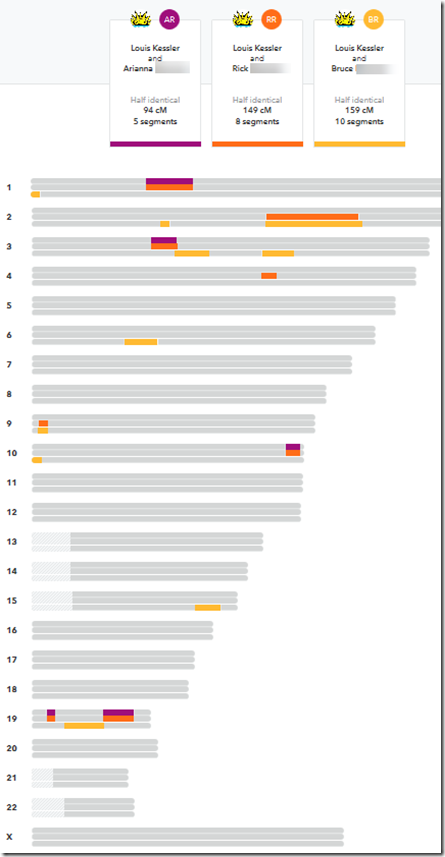
Evert-Jan and I discussed this for a while. Why were Bruce and his niece Arianna not triangulating with me? I then went to 23andMe and compared our shared DNA in their chromosome browser:
Sure enough. The 5 segments that I share with Arianna, Bruce does not share with me. Arianna, Bruce and I do not triangulate.
But it’s even worse. Bruce and his brother Rick only share the same segment with me in two places, the large segment on Chromosome 2, and the very small one on Chromosome 9. It could have been that Bruce and Rick might not even have shared those same two segments with me. In that case, Bruce, Rick and myself may not have triangulated. Then we’d have a case where two brothers would never be put in the same cluster using triangulation groups.
Evert-Jan summed it up: He said to me: ”TG clustering breaks up family bonds … and if you were using ICW they would most probably be placed in the same clusters.”
Some Thoughts
I think the conclusion is that triangulated segments and triangulated groups give good information to help you try to determine who might be your common ancestors. But they are not all-inclusive and close relatives need not have been passed down the same segment of DNA that the rest of the group received. Therefore using triangulated segments may separate close relatives when clustering.
Whereas ICW (In Common With) information will never separate close relatives and therefore is likely better for cluster analysis than triangulated groups.
Very interesting. Not every analytical technique works out exactly as expected.
Evert-Jan was initially wondering if anyone wrote about this before. I told him I that other than Jim Bartlett’s article, I doubt it, because Evert-Jan’s clustering program is the first tool ever invented that has let us look at clustering of triangulation groups.
So now it will take some thinking. Despite the possible breakup of close relations, can the ideas of triangulation group clustering still be used? Is there maybe some way of merging the ICW cluster information together with the triangulation information? We don’t know yet, but great analytical minds like Evert-Jan will be thinking hard, and that will ultimately result in new ideas along with new and better analysis software for you to analyze your DNA matches.
Update: March 21:
I noted that the TG clustering did split up a family into 2 clusters which is a problem. But I failed to mention that the 4 clusters are still good clusters, where the people in each cluster do appear to be from the same family.
Since shared match clustering gave me one big cluster because of endogamy, I didn’t get anything at all from that. But Evert-Jan’s TG clustering gave me 4 clusters, provided me information where shared match clustering alone did not.
Maybe Evert-Jan can figure a way to parlay the information that the TG clusters have to make the shared match clustering even better.


Joined: Sun, 24 Mar 2019
1 blog comment, 0 forum posts
Posted: Sun, 24 Mar 2019
Hi Louis, I’ve written about it before (and you even commented on my post!). http://twigsofyore.blogspot.com/2018/03/triangulation-is-icing-not-cake.html.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sat, 6 Apr 2019
Ah, yes. Evert-Jan: Take a look at Shelley’s article.