When I was first learning about autosomal DNA analysis, just over a year ago, I was under the misconception that if three people triangulate on a segment, then that segment is IBD (Identical by Descent).
A segment that is IBD is one that is passed down from a common ancestor. These are the segments you are looking for. By identifying people who received the same IBD segment, you can use traditional genealogical research to trace back and figure out who was the common ancestor. This is way autosomal DNA analysis and genealogical research can work together to extend your family.
Triangulation means that Person A matches Person C on the segment, Person B matches Person C on the segment AND Person A matches Person B on the segment. If other people also triangulate with them over the same segment, then all people must match all the other people on the segment. Under this condition, all the matching people are said to be in the same triangulation group for this segment. If the segment is IBD, then they all share a common ancestor who has passed them that segment or some portion of that segment.
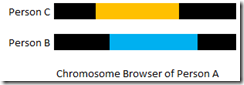
In a Chromosome Browser, if you are Person A, what you see is:
So you as Person A match Person C, and you as Person A also match Person B over the same segment. Don’t make the mistake of thinking that this means you triangulate with Persons B and C. You still have one more check to make. You must verify that Person B matches with Person C on that segment before you can say that the three of you triangulate. Unfortunately, your own Chromosome Browser will not tell you that.
Simply using what’s called ICW (In Common With) is not good enough. That means Person B and Person C are considered a match with each other. They definitely match on some segments, but those matches might not be the one specific segment you are looking at.
If you are at GEDmatch, you can do a one-to-one check of Person B against Person C and see if they match on the segment. At Family Tree DNA, you must ask either Person B or Person C to check in their Chromosome Browser to see if they match each other on the segment. At 23andMe, the “Y” indicator will tell you if you and a second person triangulate on at least one segment with a third person and then you can set one of them as the primary person in their Chromosome Browser and see if they match each other on the segment.
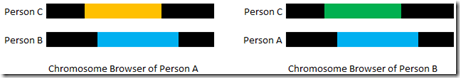
A Chromosome Browser does what I call “Single Matching”. It compares one person to all that person’s matches. By comparison, “Double Matching” uses the matches of two people and combines them and doing so can find all the triangulations that those two people have between them. It’s like having two Chromosome Browsers side-by-side:
Using double matching, you can tell that Person A matches Person C (orange) and that Person B matches Person C (green) and that Person A matches Person B (blue) all on the same segment. This is what Double Match Triangulator does for you.
That’s how you can find if people triangulate on a segment. Now what can you say if you have a triangulating segment?
My misconception a year ago was thinking that segments that triangulate are all Identical by Descent and thus the People who triangulate must share the common ancestor who passed the segment down. Debbie Kennett took a fair bit of her personal time back then through a number of emails and explained to me that this was not necessarily true. Thank you, Debbie, for your time and patience. My misconception has been corrected.
Here are the correct rules and what you need to know:
All IBD (Identical by Descent) segments will triangulate.
but
Some segments that Triangulate are not IBD
This is very important, especially the 2nd statement. Never assume that a triangulation is IBD. Here are a couple of reason why they may not be:
1. One of the People Match by Chance
A matching segment in a Chromosome Browser means that we half-match. Each line the Chromosome Browser shows actually represents a pair of chromosomes, one from your father and one from your mother. To be IBD, one of your parental chromosomes must match just one of Person B’s parental chromosomes and just one of Person C’s parental chromosomes. What can happen instead is that they match by “zigzagging back and forth” between the parental chromosomes. Check out the section “False Positive or Identical by Chance Match” in the article Concepts – Segment Size, Legitimate and False Matches by Roberta Estes who explains this very well.
You could theoretically have 3 people who all match each other zigzagging back and forth between their parental chromosomes. But this is quite unlikely, since instead of just one chance match, for this you need three chance matches. I haven’t seen any studies of this but my reckoning would be that it would be a rare event.
Still, there’s a much more likely way for a triangulation to happen with a chance match. Let’s say a segment of Person A matches Person B the correct way: over just one of their parental chromosomes. Then that segment of DNA is effectively the same for both of them (only differing because of a few no-calls, misreads and/or mutations). Person C may triangulate with them on this segment by zigzagging between its parental chromosomes to match at each location. If Person C matches by chance to Person A on this segment, then Person C will also match by chance to Person B. This appears to be a triangulation but is a match by chance and is therefore not IBD.
This match by chance in a triangulation is bad because it is difficult to discern the people who are by chance matching into a triangulation group. The by chance match will match everyone else in the triangulation group. The only way to tell the difference would be to look and compare the raw data – not a simple task, and something you can’t do at GEDmatch because they don’t give you access to the raw data.
One thing triangulation does do for you is reduce the likelihood of a by chance match. In Single Matching, any match between two people that is less than about 15 cM may be a match by chance. As the segment gets smaller, the likelihood of a match by chance increases. But in triangulation, we do have one leg matching two of the people on one of their parent’s chromosome. The by chance match can zigzag on its own pair of chromosomes, but cannot zigzag on the other connection.
That forced connection reduces the worry of matches by chance but do not eliminate it. Jim Bartlett has concluded that all triangulations of 7 cM or more cannot be chance matches. He suspects that threshold might even be as low as 5 cM.
So triangulations could still include a match by chance. Be wary of triangulations under 7 cM.
2. Matching Segments are on Different Parental Chromosomes
This one really caught me off guard. I think it was in January during a discussion about triangulation on Facebook in a DNA discussion group, Blaine Bettinger posed the question to me: What if the three matches of the triangulation are on different parental chromosomes?
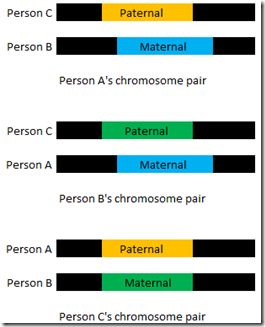
That threw me for a loop. I had never thought of that before. Blaine was right. It could happen. Let’s say person A matches person C on one specific segment on his father’s chromosome, but matches person B on the same segment on his mother’s chromosome. Person B and C would likewise have to match the same way, on the opposite parental chromosome than they do with Person A.
In the above diagram, all three people match each other on the same segment. This is a Triangulation. Person A matches Person B on the same segment (albeit different parental chromosome) as Person A’s match with Person C and person B’s match with Person C.
When Blaine first brought this up, I had a bit of a panic attack. My entry of Double Match Triangulator in the Innovator Showdown at RootsTech was coming up. This was at the time a new technology based on triangulation … and if we’ve just discovered that triangulation didn’t work, then what?
Fortunately, this turned out not to be a concern. This was just another case like the match by chance situation, where a triangulation occurs but the segment is not Identical by Descent. It does not invalidate triangulation as a tool, and triangulation is still required (but is not sufficient) for a segment to be Identical by Descent.
Don’t think these opposite parent triangulations are a rare thing. They likely are not. I’ve found a number of examples in some of the work I’ve done. And here’s the bad thing about them. It could happen with segments of any length. There is no protection by using only large segments, say 15 cM or more.
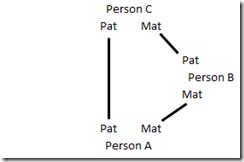
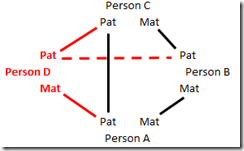
So then I thought of how to eliminate this possibility of elimination of opposite parent triangulations. Redrawing the Person A, B and C matches as a diagram, we have this:
To prevent this, all we need is a 4th person who is part of the triangulation group. When you add that 4th person, and try to connect them to each of the other three, as below:
you’ll see there is no possible way that you can make that last connection (the dashed red line) without forcing each line to be an identical segment. For example, in the above diagram, we can logic that:
A Pat = C Pat
A Mat = B Mat
B Pat = C Mat
and
D Mat = A Pat = C Pat = D Pat
so D’s parental segments would match each other,
and
C Pat = D Pat = B Pat = C Mat
so C’s parental segments would match each other,
and
C Pat = C Mat = D Pat = D Mat = A Pat = B Pat
so all people match on an identical segment
IOW, to force the triangulating segments to match, we need 4 people in the triangulation group. This could be done with an extension to Double Match Triangulation that could awkwardly be called: Triple Match Quadrangulation, which would use the segment matches of 3 people (Person A, B and C) to find all the people D who match A, B and C where A, B and C also match each other.
An alternative that would be simpler is to do two sets of Double Match Triangulations (A with B, and A with C) and combine the results. Doing so would avoid the need to ever have to use the word “quadrangulation”.
One caveat with regards to this. If any of the four people are siblings or other forms of double relatives that fully match on the segment on both chromosomes with one of the other people, then you can still include them in the triangulation group, but you can’t count them as one of the four and you’ll have to add another person who triangulates to bring the total up to four or more.
Double Match Triangulator already allows two or more sets of Double Matches to be combined in a “By Chromosome” run, but it does not yet merge the triangulation groups together. I’ll attempt to do so in a future version of DMT using this new knowledge that a 4th person in the group can guarantee that its the same segment that’s being triangulated.
Remember: Triangulation is still Very Useful
Even though triangulations may match by chance, or may triangulate on opposite parental chromosomes, that only means that triangulations are not necessarily Identical by Descent.
Despite this, what triangulating does for you is remove from your consideration a very large number of single matches that cannot be IBD. The matches that remain that triangulate are the only ones from which IBD segments will be found.
Said another way: Identical by Descent segments must triangulate, so by triangulating you are greatly reducing the matches you have to look through to find the common ancestors of your DNA relatives.
Just be wary. Don’t assume all your triangulations are IBD.
—-
Update Sept 25: Ann Turner informed me on the ISOGG Facebook group that 23andMe allows you to put anyone as the primary person in the Chromosome Browser, so you can determine by yourself if Person B matches Person C without having to contact them. I’ve now corrected that in my article.
Additional notes in my reply to Ann:
From a mathematical analysis, I am pretty sure you would only need 3 people’s matches, A, B & C to triangulate an unlimited number of people: D, E, F, G, …. into the same triangulation group (which is what I horribly named: Triple Match Quadrangulation). Using only two people, A & B, will determine all the triangulations, but those might include some triangulations with matching segments on different paternal chromosomes.
So no, you wouldn’t need to compare D & E. If you add F, you wouldn’t need to compare D & E, D & F or E & F. Etc.
What you will find, however, is that some people may not have enough total cM to make a match criteria. They may match just on a few small segments including the one of interest. But you can’t tell because they are not an overall match for you. You will then need to match them on the segment with a third person from the triangulation group to prove that they belong.

Joined: Mon, 25 Sep 2017
5 blog comments, 0 forum posts
Posted: Mon, 25 Sep 2017
Quick note: At 23andMe, you don’t need to ask one of the matches to confirm that they share a segment in the same place. You can compare them directly to one another from your own account.
Joined: Mon, 25 Sep 2017
5 blog comments, 0 forum posts
Posted: Mon, 25 Sep 2017
There is a third possibility: Triangulated segments could be identical by descent, but not by descent from a recent common ancestor. This most often happens in endogamous populations, where individuals are related to one another multiple ways. For example, you and a match may have identified your most recent common ancestor couple as Mr A and Ms B, but you might also be related through Mr C and Ms D, Mr E and Ms F, and Mr G and Ms H. You may find a third person who truly triangulates with both of you, but you won’t know which ancestor passed along that segment.
This latter issues is especially problematic when one or more people in the triangulating group do not have trees that are filled in for many generations. That is, there may really be multiple connections, but you can’t tell because the tree doesn’t go back that far. It’s easy to assume that you’ve proven that you share, say, a 4th great grandparent when really the shared segment came from a more distant ancestor.
Finally, even without recent endogamy, you can get apparent triangulation groups that pre-date genealogical time. The shared ancestor may have been 1000 years ago and have left so many descendants that the segment in question was present in a large percentage of the population 200 years ago. This is called “excess IBD” in the scientific literature and “pileups” in common terms. In the case of a pileup, a triangulation group does not help you identify the common ancestor.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Mon, 25 Sep 2017
thednageek: I think Ann beat you by a couple of minutes regarding 23andMe, so I had to give her credit for that update.
Re your 3rd possibility: Whether a segment is from a recent common ancestor or a distant common ancestor, it’s still the same segment from the same ancestor that all the people in the triangulation group have in common. Mathematically everything is correct and the triangulation is doing its job as stated.
So then, as you state, the problem is in using this information to help you when the ancestor is too far back. I think we can come up with techniques to help figure that out, based on the idea that triangulation groups with a distant common ancestor often will be contained in a larger segment having a triangulation group with fewer people from a closer common ancestor. One of the techniques I’m looking at is to start big (as far as segments go) with the largest triangulations and work your way back in time by finding smaller segments with triangulations of additional people.
Joined: Mon, 25 Sep 2017
5 blog comments, 0 forum posts
Posted: Tue, 26 Sep 2017
Louis, you said “One of the techniques I’m looking at is to start big (as far as segments go) with the largest triangulations and work your way back in time by finding smaller segments with triangulations of additional people.”
I’m curious how successful you’ve been with that. Yes, a “pileup” segment might be embedded within a larger segment that triangulates with a close relative, letting you, say, assign the connection as being through a specific set of great grandparents. But where do you go from there? Do you have enough known relatives who match on that segment to continue narrowing it down? And if you do, how do you prove that those matches inherited the segment in question from the MRCA?
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Tue, 26 Sep 2017
Thednageek (Leah):
Excellent questions, and thank you for them because you’re making me really think. I’m hoping to use triangulations to incrementally build up a chromosome map of ancestors the way Jim Bartlett has done and describes on his segmentology.org blog. It is still a work in progress. From a mathematical perspective, every extra person whose segments match with you adds valuable data to help develop the map. I believe there’s more than enough data to make it possible to be programmed. The key is to start with yourself and your segments and work your way back, identifying parental segments, grandparent segments, etc., as far as you can go.
Obviously, you can only go as far back as you’ve genealogically researched. Once you get to, say a set of great grandparents who are your dead end, you could be stuck. You may be able to go further back analytically, but if so, it will just be a blind connection no better than knowing you’re a DNA cousin with someone.
“Proving” a distant MRCA is likely seldom possible. You gave an excellent example on Facebook of being descended from one early Acadian in at least 26 different ways. Are you sure he was the MRCA? Maybe it was his grandfather, or granddaughter, or that segment didn’t come from him at all. Triangulation doesn’t prove anything. All it does is filter out non-IBD segments to give you a better chance to find the IBD ones.
I think trying to use triangulation to identify a common ancestor has many problems. Using triangulation to build up a chromosome map of your ancestors is a more worthwhile task to pursue.
Joined: Wed, 27 Sep 2017
6 blog comments, 0 forum posts
Posted: Wed, 27 Sep 2017
I presume you are using triangulation here to mean that three people match on the same segment and that close relatives (eg, parents) cannot be included in the triangulation as they are on the same leg of the triangle as you. If so, it is not true to say that three people have to match on a segment for it to be IBD. It’s actually very hard to get three people to match on the same IBD segment. For all the segments where I’ve been able to identify a recent genealogical common ancestor I only have a simple pairwise match (excluding my parents).
I agree with you and Leah that using triangulation to identify a common ancestor is likely to be problematic for the reasons that Leah has explained. However, I think you’re right that triangulation is potentially useful for chromosome mapping. It’s also very helpful for identifying pile-up regions. The scientific papers on IBD detection show that there is a direct link between haplotype frequency and the age of a segment, so if lots of people match on a segment it is much more likely to be ancient than recent.
There is another possibility to consider to explain these triangulated segments. It’s quite possible that they are identical by state but not identical by descent. In other words, even if you can correctly assign the alleles to your two parental chromosomes through phasing, we can still expect some false coincidental matches. This is because we are not sequencing the whole genome and we are only inferring segments. We don’t know what SNPs are missing from the inferred segments, but if we were able to sequence all the SNPs in our genome some of these segments would turn out not to be real after all. This could occur in regions of the genome where there is low SNP density or where the alleles within the segments are widely shared in the ancestral population. We’ve seen this effect at AncestryDNA where, even with phasing, there are still a lot of false positives (or false negatives). See my blog post here:
https://cruwys.blogspot.co.uk/2017/08/comparing-parent-and-child-matches-at.html
This again is mostly a problem with smaller segments under 15 cMs. In my experience, it’s only the segments under 15 cMs that triangulate. It’s also noticeable that these triangulated segments are mostly in pile-ups. A lot of my triangulated segments seem to have very few SNPs.
I wrote a blog post a while back now with some hypotheses to explain triangulated segments:
https://cruwys.blogspot.co.uk/2016/01/autosomal-dna-triangulation-part-2.html
I would love to be able to find a way to test some of these different hypotheses but I don’t think we’ll be able to do so until all the companies deliver phased whole genomes.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Wed, 27 Sep 2017
Debbie: I agree with everything you say (except maybe one thing, which I’ll likely write about in a future blog post).
I was specifically talking about triangulation. Yes of course, a match between two people can be IBD. But the point of the article was to dispense with the notion that triangulation means IBD, as you taught me a year ago.
Yes, inference can also result in false positives. My mathematical and statistical background makes me a big hater of inference. And the DNA industry seems to be getting deeper into that all the time. Soon a segment match won’t mean anything at all. My reads of AncestryDNA’s white papers leave me with little confidence in what they are doing. I’ve done the same parent-and-child comparison with FamilyTreeDNA as you did with AncestryDNA data, and my results show that FamilyTreeDNA gives significantly fewer false positives above 6 cM: http://www.beholdgenealogy.com/blog/?p=2003
I deal with endogamy. It seems like I have different observations than you do with your data. I match with 11,400 people and I have lots of triangulations. For example, I use my Double Match Triangulator program to compare my segment matches (my Chromosome Browser Results file) one by one with each of the 32 Chromosome Browser Results files that I obtained from people that I match with, none of whom are closer than a 3rd cousin. Comparing my file with one of those 32 files gives on average 3625 triangulations with 2344 people. Put together, that gives me over 100,000 triangulations to play with.
But yes as you say, very few of these triangulations are within a genealogical timeframe which for me is only 5 generations. My thousands of triangulations are not just within certain pileup regions, but are everywhere. They encompass almost every cM of every chromosome. I am going to see if I can use all this mass of triangulations to put together my chromosome map that extends back at least a few generations and possibly even back to my 5 generation horizon. But it will take some ingenuity and programming as well as a bit of trial and error to do so.
Joined: Wed, 27 Sep 2017
6 blog comments, 0 forum posts
Posted: Tue, 3 Oct 2017
Louis, I shall look forward to your future blog post.
Inference is a popular word in population genetics and genetic genealogy. Unlike mathematics, we can’t have complete certainty. It’s therefore a question of testing hypotheses and quantifying the uncertainty. I’ve actually been very impressed by Ancestry’s white papers and their transparency about their methodology. I wish FTDNA would do the same.
Parent and child comparisons at FTDNA can’t be directly compared with those at AncestryDNA. Each company has different match thresholds:
https://isogg.org/wiki/Autosomal_DNA_match_thresholds
As you will see, FTDNA won’t report a match unless there is a single segment of at least 9 cMs. If they were to report all segments down to 6 cMs as Ancestry do, then I would expect to see a much higher false-positive/false negative rate because of the lack of phasing.
It would be interesting to see comparisons of distributions of triangulated segments from people who descend from different populations. I think that the more endogamy there is in a population the more we would expect to see people triangulating on lots of segments. The difficulty is that with endogamy you will have multiple possible pathways by which the segments could have reached and no way of determining the correct route. I believe you will be able to use these segments for chromosome mapping. I suspect you will have to wait until we are able to use whole genome sequencing for more accurate relationship predictions. WGS will allow us to drill down into the rare SNPs within the segments.
The difficulty I have with the small triangulated segments is that nearly all the people in my TGs are Americans. They don’t have any shared surnames in common with me and no identifiable locations outside of the US. I don’t have any ancestors who emigrated to the US and left descendants. This rather suggests that these segments, if they are real, must all be very ancient, and probably predate the colonisation of America.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Tue, 3 Oct 2017
Very interesting, Debbie. So then it seems that FTDNA’s criteria of at least one 9 cM segment is better (i.e. produces fewer false matches) than AncestryDNA’s combination of phasing and inclusion of segments down to 6 cM. That may be indicative that AncestryDNA’s phasing combined with its Timber may not be doing as good a job as it is claiming to be doing.
Joined: Wed, 27 Sep 2017
6 blog comments, 0 forum posts
Posted: Wed, 4 Oct 2017
No I think it means the opposite. FTDNA can’t use a higher threshold because they are not using phasing or any algorithms to remove pile-ups. The vast majority of matches that don’t match either parent at AncestryDNA share under 9 cMs. Ancestry’s own tests published in their white paper show that we would expect some mismatches. See this previous thread in the Genetic Genealogy Tips and Techniques group: https://www.facebook.com/groups/geneticgenealogytipsandtechniques/permalink/314023089061398/ The matches we get at Ancestry are more reliable than those we get at FTDNA.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Wed, 4 Oct 2017
Debbie: Well, I’m just looking at the results. And FamilyTreeDNA seems to have many fewer disprovals of IBD by parents at any given cM level than AncestryDNA does.
Joined: Sun, 18 Feb 2018
3 blog comments, 0 forum posts
Posted: Sun, 18 Feb 2018
I find the following GETMAtch 1-1 matching segments among known but distant American cousins (and my brother) and a potential English cousin (16 degrees between me and the English cousin for instance). I wonder if Chr 1 would indicate a shared IBD this many generations back? Due to Chr 15 pileup, I suspect those can be immediately discounted. Oral family history suggests as much but there is no documented evidence. I would welcome opinions!
1 742,584 2,017,761 4.1 115
1 742,584 2,296,555 5.1 182
1 742,584 2,108,199 4.4 127
1 742,584 2,296,555 5.1 183
15 25,312,725 27,216,380 5.6 235
15 25,517,776 27,229,496 4.7 218
15 25,568,286 27,679,734 5.4 295
15 25,582,763 27,491,292 5 227
15 25,582,763 27,214,074 4.4 192
15 25,645,234 27,214,074 4.2 107
15 25,650,782 27,214,074 4.2 168
15 25,687,195 27,884,370 5.2 280
15 25,687,195 27,214,074 4 153
18 6,439,309 7,365,434 4.8 180
18 6,652,650 7,845,868 5.9 304
18 6,771,016 7,679,332 4.6 278
18 6,871,677 7,747,048 4.4 267
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 18 Feb 2018
tewilso:
What I can’t tell from your example is who is matching who.
Unfortunately, and especially for smaller segments under 7 cM, chance matches can happen because the companies are comparing both parental chromosomes of one person with both parental chromosomes of another chromosome. And that is at every SNP so one SNP may match father to father, the next father to mother and the third mother to father. Those would all be called a consecutive match of 3 SNPs since they cannot be told apart.
Because of this, for matches of segments under 15 cM you need more evidence to show that they may be IBD. First of all, every person matching on the segment must match everyone else. So first you must use GEDmatch 1-1 on Person A with B, person A with C, person A with D, etc., and then Person B with C, Person B with D, etc., and the Person C with D, etc. Anyone who does not match with everyone else is not a part of the same triangulation group.
Once you get people that you have all triangulating with each other on a segment, then you MIGHT have an IBD segment. That’s not guaranteed. For small segments, you still might have some of the people matching by chance as my article states. At this point you can try to see if you could figure out who the common ancestor might be that could have passed down that segment to all of them.
There are a lot of “coulds” and “mights” in there. The bottom line is that the DNA matching evidence must work together with your genealogical research, and if they both support each other, then it gives you further evidence to help support your hypothesis that you all share an IBD segment from a specific common ancestor.
Sorry there’s no simple yes or no answer for you.
Louis
Joined: Sun, 18 Feb 2018
3 blog comments, 0 forum posts
Posted: Sun, 25 Feb 2018
Update: By analyzing all pairs, I have two cousins that triangulate (GEDMatch one-to-one comparisons) with a potential cousin on Chr 1 (all separate pairs match on this same segment.) The known cousins are my 5th-6th cousins, the potential cousin would be our eighth cousin. Ditto on another segment (Chr 18) but with two different known cousins and the same potential 8th cousin. Due to the large degree separation to the potential 8th cousin, the SNPs are ~200, 300 and at 4.2 and 5.2 cM on the two different triangulated segments , Ch1 and 18.. What is the confidence level that we share common ancestor? Still by chance? Are false positives quantifiable so one could ascertain the probability of common ancestor?
Joined: Sun, 18 Feb 2018
3 blog comments, 0 forum posts
Posted: Sun, 25 Feb 2018
We have a very good idea of who the common ancestor may have been - Lt. Gen. Emanuel Scrope Howe. His father was 9 degrees above Princess Diana.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Thu, 1 Mar 2018
tewilson: Well, DNA and genealogy together can lead you to reasonable conclusions as long as it all makes sense. So just make sure you’ve got all your i’s dotted and your t’s crossed.