Over at the ISOGG Facebook group, Rich Capen asked if it would be possible for Double Match Triangulator to compare GEDmatch kits. Well, that spurred my interest to see if I could.
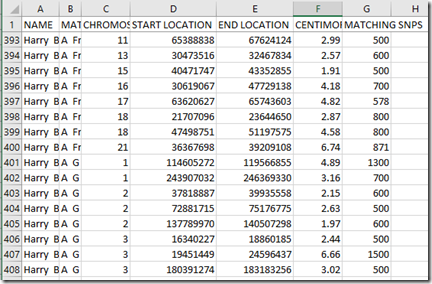
My uncle’s Chromosome Browser Results (CBR) file at FamilyTreeDNA is very big because he is Ashkenazi. I just downloaded a new one so I can compare with GEDmatch. The first 3 attempts failed and it didn’t download the whole thing. The 4th finally worked and I downloaded the file. After selecting Download All Matches link at FamilyTreeDNA, I had to wait 30 seconds or so before anything would happen. Then the browser’s Save File box would pop up. Patience is a necessity here. My uncle currently matches with 9,288 people at FamilyTreeDNA. (Still, only one of them is a confirmed relative). The Chromosome Browser Results file that was downloaded contains 203,035 individual segment files for these people, an average of 22 segment matches per person.
The CBR file looks like this:
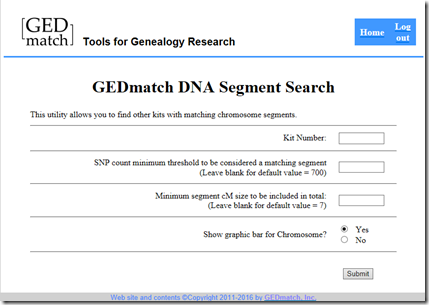
I then went over and gave a $10 donation to GEDmatch (which is less than what they should charge) so that I could get access to their Tier 1 utilities to get the segment matches that they found:
I entered my uncle’s GEDmatch kit number. FamilyTreeDNA downloads all segment matches down to 1 cM. GEDmatch has a default minimum of 7 cM. I entered the minimum GEDmatch would allow (5 cM and 500 SNPs) and I pressed the submit button.
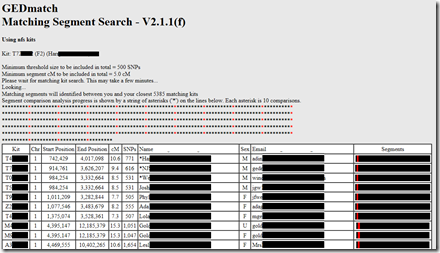
GEDMatch showed a progress screen and after a couple of minutes finished processing and displayed the results in the browser Window. It said that matching segments will be identified with the closest 5,385 matching kits.
Right off the bat, I do like the Sex column, because if it is specified, then I don’t have to use the segments to guess at the sex (and I’m not even sure if that’s even possible yet, but it’s something I’m I will eventually want to do).
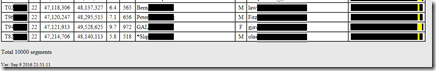
At the bottom of the file, it said: “Total 10000 segments”. That got me worried since it’s such a round number. Does GEDmatch have a limit of 10,000 segments that it will give you?
Well, the listing does go down to Chromosome 22, location 49,528,625. The FamilyTreeDNA CBR file goes to Chromosome 22, location 45,772,802. I would guess that GEDmatch is listing everything up to Chromosome 22 and maybe the result of getting exactly 10,000 was just a fluke.
However, the Chromosome 23 (X matches) are not included in the GEDmatch listing. It ends at 22. That’s not good. FamilyTreeDNA’s CBR file includes Chromosome X.
Okay then. Let’s put the GEDmatch data into a spreadsheet. GEDmatch does not download a file. The only way to do it is to select all the data on the browser page, copy it to the clipboard, and paste it into Excel. But 10,000 rows in my browser was too many to select. It wouldn’t copy to the clipboard. I found if I selected a few chromosomes at a time, it would copy and paste, so I did it in about 8 steps, and eventually loaded the spreadsheet with the 10,000 match lines and the one header line.
Next step: Let’s see if we can compare the data in the two files. Is it compatible? To do so, I need to only include GEDmatch data that’s from FamilyTreeDNA.
The 10,000 matches are for 1,853 kits. That means there’s only an average of 5 segment matches per kit (person) and that’s because of the 5 cM minimum match length versus FamilyTreeDNA’s 1 cM minimum which gives 22 segments per match.
Of the kits, 562 are prefixed by A, meaning they’re from AncestryDNA so I’ll get rid of those 3,193 matches. 559 are prefixed by M, meaning they’re from 23andMe, so I’ll get rid of those 2,466 matches. There are 3 kits prefixed with W having 3 matches and 32 kits prefixed by Z with 174 matches. I’m not exactly sure what those are, but they’re not FamilyTreeDNA, so I’ll get rid of them too.
That leaves me with 698 matching kits prefixed by T which are the FamilyTreeDNA kits. They total 4,164 matches and I’ll keep them.
Now for the next problem. The person’s name listed in the GEDmatch data is quite often not the same as the person listed in the FamilyTreeDNA CBR file. The CBR file does not have an email address in it, so that can’t be used for verification. But there are some that match. I can look at those.
665 names in GEDmatch were not matches in my FamilyTreeDNA file. That meant that only 34 names were the same. They have 208 matching segments in GEDmatch (down to 5 cM) and 753 matching segments in FamilyTreeDNA (down to 1 cM).
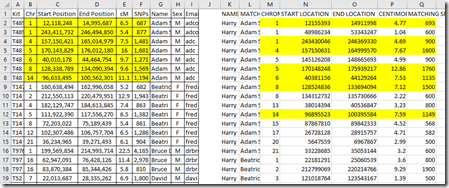
Now it becomes clear. The bad news. GEDmatch and FamilyTreeDNA do not give the same Start and End locations for the matching segments, nor are the cM or SNPs the same. See a comparison of the first person below. I’ve highlighted the matching segments.
I am very disappointed. What this means is that GEDmatch results cannot be combined with FamilyTreeDNA matches. Double Match Triangulator will not be able to use them both together.
But would it be possible for me to modify to Double Match Triangulator to be able to work with GEDmatch matches? Well technically yes, I could. But to me it’s not worth the effort. There are too many things to check to make sure the file is correct.
So Can DMT be Used with GEDmatch data?
Yes. If you want to use DMT with your GEDmatch Tier 1 segment matches, then this is what you can do. It should only take you 5 minutes or so if you know Excel well enough:
- Login to GEDmatch. You will need to be subscribed to the Tier 1 utilities. If you are not, you can subscribe for a month for $10.
- In the Tier 1 Utilities, click on “Matching Segment Search”. Select “No” for “Show graphic bar for Chromosome?”, and either submit that with the default settings, or lower SNP to 500 and cM to 5 (their minimums).
- Copy the displayed GEDmatch match table with the table headings, and paste them into a spreadsheet like columns A to I in the diagram above. Note: Internet Explorer and Microsoft Edge may be unable to copy the table if it is very large. Google Chrome, Firefox and Safari seem to work for any size.
- In the spreadsheet you created, in columns K to Q, row 1, put the column headings that a CBR file has, which are:
NAME
MATCHNAME
CHROMOSOME
START LOCATION
END LOCATION
CENTIMORGANS
MATCHING SNPS - In column K, put the name of the Kit owner
- Copy the Name Column G to the Matchname Column L
- Copy Chromosome, Start and End Locations, cM and SNPs from Column B to F and use “Paste Values” to put them into columns M to Q. That will get rid of the comma format. Alternatively, you can format those columns as “General” which will get rid of the commas.
- Delete columns A to J.
- Save the file as a CSV (Comma Delimited) file and give it a file name typical for a CBR file starting with the Kit number, e.g.: A123456_Chromosome_Browser_Results_20161103.csv
DMT should be able to input that file.
Of course, what you now need is a second one of those files from another kit at GEDmatch. GEDmatch lets you work with any kit that is visible to you, so all you need is the kit number.
Note that GEDmatch’s Tier 1 segment matches exclude people who match more than 2100 cM. They say they are doing this so as not to obscure the matches that you’re really looking for. This means matches with parents, children, and siblings are left out of the Tier 1 match results. If you want them included, you’ll have to run GEDmatch’s one-to-one match and manually add their matches to the file.
One Little Trick
If you make a copy of the file, you can run DMT using the file for Person a and the copy as Person b. Every segment match will double match but you’ll end of with a nice Map of the matches and and a People page listing all your matches.
You will still be stuck with the GEDmatch limitations
- Minimum 5 cM, 500 SNP matches
- Possibly 10,000 match limit.
- Possibly no X matches.
- Cannot mix and match GEDmatch and FamilyTreeDNA CBR files.
But it should still give you lots of new double matches to keep you very busy.
Update: Dave Sherry on the ISOGG Facebook page said that FamilyTreeDNA uses Build 37 and GEDmatch uses Build 36, hence the differences. GEDmatch will at some time in the future, have to convert to Build 37 for the locations to be the same. Or maybe there is a utility out there to convert base addresses from Build 36 to Build 37.
Update Apr 28, 2017: I improved the steps (now 9 of them) to convert the GEDmatch data to work with DMT. I’m going to try to add direct support of GEDmatch format into the next version of DMT so that this manual conversion will no longer be necessary. Also want to do the same for 23andMe match files.

Joined: Thu, 17 Nov 2016
1 blog comment, 0 forum posts
Posted: Thu, 17 Nov 2016
Hello–got to your site via a link from Jim Bartlett’s blog. What I’m looking for is software that enables me to create a chromosome map, matching segments to ancestors. Is this what DMT does? Thanks.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Thu, 17 Nov 2016
Currently what DMT does is to produce a chromosome map, showing double matching segments. So you need to give it two Chromosome Browser Results files - one for the primary person, and one for a match of that person to compare with.
No, it does not yet match to ancestors. The ultimate goal is to get it DMT to use that by using crossover boundaries and Double Match Groups to map the likely ancestors on each segment. Of course, the ancestors will be identified by a possible location in an ancestral tree but will be nameless. This will take a fair bit more analysis and experimentation to come up with the methodologies needed. In the meantime, at least, DMT will produce the Double Match information making it easier for you to do this analysis yourself.
Louis