The IAJGS Conference 2017 - Mon, 31 Jul 2017
I’ve returned from Orlando after the week-long #IAJGS2017 (International Association of Jewish Genealogical Societies) Conference on Jewish Genealogy. This was my 8th International genealogy conference, my first being RootsTech 2012, but it was my first IAJGS Conference.

The venue was The Swan Resort, ideally situated midway and within walking distance of Epcot and Hollywood Studios at Disney World, Florida. A mid-July visit to central Florida means 88F (32C) days with high humidity and thunderstorms possible at any time. So the hotel and Conference Centre compensate with powerful air conditioning that dresses up attendees in long sleeves, slacks, and even sweaters and jackets.
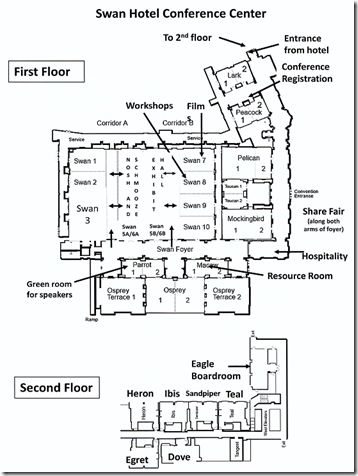
The Conference facility was just the right size to handle the 900 attendees, 9 different tracks, an exhibit hall, a computer workshop room, a resource room with computers giving access to paid services for free, and a mentors area. It was in a separate wing of the hotel, so the conference attendees had the whole area to ourselves.
Sunday
I arrived late Saturday night, so I picked up my Conference Package Sunday morning. I loved the name tags, which prominently displayed up to 5 ancestral surnames and places. It was a no-brainer to compare them whenever meeting someone for the fist time, or for the tenth time. I added my own ribbons at the bottom and I got the ProGen ribbon at our Thursday BOF meeting. The penguin sticker was from Banai Feldstein and I’ll say more about that below.
The Conference had an App the same as the RootsTech App. You could plan the events you wanted to go to, view the Twitter #iajgs2017 feed, see the list of speakers and attendees, invite them to be friends, view all the handouts, etc. Since I have a Windows Phone, I had to use the online version with wifi. The wifi was good in the main areas, but was a bit spotty in some of the conference rooms. Due to extensive use during the conference, my phone battery would typically run out midday. so I’d then whip out my backup phone and start with that. By Wednesday, I started carrying a charging cable and plug in my back pocket.
Sunday started with a New Attendee Welcome. As this was my first IAJGS, I decided to go to it. Two of the conference co-chairs, Dennis Rice and Adam Brown presented and led the discussion.
Arriving late about 9:30 a.m. I heard the end of Shelley Talalay Dardashti talk describing DNA matching at MyHeritage:
At10:30, I listened to Bennett Greenspan tell us about Family Tree DNA. My notes from his talk included him saying: “There will be some new tools coming out in the next quarter or two, like triangulation.” Also of interest to me, he said “If you have 4 Ashkenazi grandparents, you’ll have 12,000 matches matching to 3/4 of all Ashkenazi Jews who have tested.” I was always curious about this. Bennett’s statistics implies there are about 16,000 Ashkenazi who have tested at Family Tree DNA. That implies that a 100% Ashkenazi with 12,000 matches should have a Pickholtz-Diamond Index at Family Tree DNA of about 20. I also wrote this in my notes: How often do you get the owner of a company talking with such passion about a topic?
At 1:30 pm was a presentation I was really looking forward to. My mother’s mother comes from a family of 17, and I had always heard that 14 of them were killed towards the end of WWI. I found that odd. What was going on in Ukraine during WWI that I didn’t know about? It was well before the Holocaust, and after most of the pogroms in Eastern Europe that prompted my ancestors to emigrate to Canada in the early 1900’s. I had never found out more about that, until now.
“My Dear Children” is a historical documentary being produced about the anti-Jewish massacres in Eastern Europe from 1917 to 1921. LeeAnn Dance, the producer of the film, spoke to us about it and presented clips from the upcoming production. As many as 250,000 Jews were wiped out in this massacre. Expected release is in October on PBS.
I took the rest of the afternoon to wander around the Exhibit Hall. It was small compared to other larger conferences I’ve been at, but all the major players were there. I took some pictures of their booths.
RootsMagic (Bruce Buzbee and his wife Laurie): 
Family Tree Maker (Jack Minsky and Mark Olson): 
In the evening at 7:30 p.m. was the first Keynote. Swan 1, 2 and 3 were combined for the Keynotes. There was quite a crowd there for each keynote session: 
Marlis Humphrey, the IAJGS President greeted the crowd and gave some interesting statistics: This conference has 900 attendees, 225 first timers (that includes me!). 200 have attended 5 IAJGS conferences. 100 have attended 10 or more. This conference has the most extensive DNA track ever at an IAJGS Conference. 10% of the attendees are from overseas. 34 states are represented. This is the first ever IAJGS conference in Florida. More than 25 attendees from Florida.
Daniel Horowitz then came out and prior to introducing the speaker, talked about MyHeritage who was the sponsor of this keynote. Of interest, I noted he said that the following were coming at MyHeritage DNA: Pedigree charts, a chromosome browser, and matches by place.
The keynote was Professor Robert Watson who gave an interesting perspective on Alexander Hamilton and how his life connected to the Jews of the Caribbean island of Nevis: 

Following the keynote, I was accosted by a couple of penguins, who had announced earlier that all 7 continents were being represented. Banai Feldstein and Ron Arons (and a number of others who I don’t have pictures of) represented the Antarctica Jewish Genealogical Society, which has a private Facebook group that they graciously allowed me to join: 

Monday
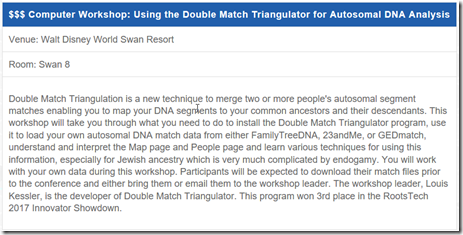
Bright and early at 8:15, was my computer workshop on Double Match Triangulation. It was in Swan 8, a conference room with chairs behind rows of tables so people had a place for their laptops or pads, and it had its own wifi. But there were no computers in the room. I had about 30 attendees and I knew prior that about 8 of them would need a computer for the workshop because DMT currently only runs on Windows. We were expecting Windows laptops to be supplied to those who needed them, but they turned out to be Chromebooks that won’t run DMT, so we had to make do.
Sandy Aaronson introduced me and helped the workshop attendees when they had problems. Sandy had assisted me early on in the testing of DMT last summer, and provided me with a lot of feedback that helped in its development. I communicated extensively with Sandy over the past year, so it was nice to finally meet her in person.
I was unexpectedly surprised to see Bennett Greenspan in the 3rd row with his computer. He hadn’t registered, but there certainly wasn’t anyone who was going to tell him he couldn’t attend. Bennett was genuinely interested in what I was doing.
It’s difficult to give a workshop when there are different people at different levels. You just have to just keep it rolling so that everyone can learn within the 90 minutes what you wanted them to know how to do. This was my first ever workshop/presentation on DMT, so it was very nice to have a half-dozen of the participants come up to me later at the conference to say how much they enjoyed my workshop.
The workshop was scheduled to run until 9:45 a.m. Sandy and I stayed another hour to answer questions and provide additional help to those who wanted.
This workshop was my only personal commitment to the conference. It was nice to be scheduled early so that I now had 3 full days to enjoy freely. There weren’t any talks for a while that I just had to see, so I took a few hours to get some lunch, walk around the Swan/Dolphin resort, walk to the Boardwalk area to the Epcot entrance, back all the way to the Hollywood Studios entrance, and back to the hotel. Ended up that day with 18,874 steps.
For the afternoon and evening, I picked a few DNA sessions. At 3:30 pm was “What Y-DNA Lineages Can Tell Us About Jewish History and Migration” with Rachel Unkefer, Adam Brown, Michael Waas and Janet Akaha on the panel.
Following the panel discussion, they asked for questions and I supplied one. I asked whether Y-DNA has discovered connections to any of the 10 lost tribes of Israel. I said that several years ago, Simcha Jacobovichi, an Israeli-Canadian historical documentary film-director, did a movie on the Quest for the Lost Tribes, where he claims to have found many of them. Adam Brown’s answer to me was that only one tribe has so far shown Y-DNA connections (I forget which one he said) and he classified the connections of the other tribes as fiction. Interesting.
The other movie of Simcha’s that I really like is The Exodus Decoded which attempts to plausibly explain the ten plagues and then there’s his claim of finding the true Mount Sinai. Great stuff! Watch them for yourself and make your own judgements.
At 5 p.m. was “The DNA of the Jewish People” given by Bennett Greenspan. This was a very interesting exposition about a fight that Bennett is fighting to help refute “The Thirteenth Tribe”, a 1976 book by Arthur Koestler claiming that Ashkenazi Jews are not descended from Judea, but are Khazars, a Turkic people. Koestler, of course, proposed this to dispute Jewish people’s claim to Palestine. Bennett is out to disprove this through DNA analysis. To summarize Bennett’s task, he used a saying of Ronald Coase: “If you torture data long enough, it will confess.”
Following Bennett’s talk, I was conversing with Karen Schlussel who I found out was a cousin of Bennett’s. Bennett came up to the two of us and said he was very interested in learning more about my double matching technique. So the three of us went out for dinner together.
Bennett had to rush back for the evening keynotes who he was introducing. The evening talk was “The Contested Origins of Ashkenazic Jews in Eastern Europe” so it was in effect a continuation of Bennett’s quest. This time, we were getting the research details that scientifically refute Koestler from the respected scholars: Dr. Alexander Beider of Paris and Dr. Harry Ostrer of New York.
Tuesday
I was up early for the Tracing the Tribe breakfast. This was a meeting of people who frequent the Facebook group for Jewish Genealogy. It has over 16,000 members. Schelly Talalay Dardashti, who is the moderator of the group, led the breakfast discussion about how to form your own Facebook groups for a specific town or surname, along with essential moderator and guideline issues.
Unfortunately, the TTT breakfast conflicted with the Blogger’s Brown Bag Breakfast which I also would have liked to attend. Emily Garber writes about the Blogger’s breakfast that I missed at the beginning of her Day 3 blog.
At 8:15 a.m., I attended the Q & A on Ashkenazi Surnames with Dr. Alexander Beider, who was one of the keynotes the night before. Dr. Beider told us that surnames can Monogenetic (only one person took the surname) and Polygenetic (multiple people took the surname). What we were all looking forward to was to write our surnames we had questions about on a slip of paper, and then he picked them up one by one and said what he knew of that surname’s origin and meaning. In my case, I asked him a pressing question I had as to why I have a couple of cases Focsaner/Zvoristeaneau and Segal/Hertzan that two brothers took different surnames. Unfortunately he didn’t have the answer for me. He generously gave us his email address in case we had further surname questions. If you haven’t already, be sure to check out Dr. Beider’s book: A Dictionary of Jewish Surnames from the Russian Empire as well as his other books. They are expensive to purchase for yourself, but you should find many of them in any major library. (Note: Dr. Beider did not push or mention any of his books during any of his talks. The mention is mine and mine alone.)
I went into the Exhibit Hall, and caught conference co-chair Adam Brown at his Avotaynu DNA booth. The Avotaynu DNA project is to foster academic studies illumination the history of the Jewish people. So I let Adam include me in the project. Then I spent some time talking to Bruce Buzbee of RootsMagic and his wife Laurie. Dick Eastman joined us for some more interesting conversation.
The rest of the day was basically what I’m calling “Ukraine Day” for me. The Ukraine SIG (Special Interest Group) had a series of talks starting at 9:45. My whole mother’s side as well as my wife’s side comes from Ukraine. I’ve never explored the Ukraine SIG before so this would be my initiation.Janette Silverman, Ukraine SIG coordinator and JewishGen’s volunteer of the year, gave the SIG 101 introduction.
Next up at 11:15 was Ukraine SIG 102 with more about the SIG brought to us Janette with the help of Joel Spector, Volunteer’s Director, and Chuck Weinstein, Towns Director.
Not skipping a beat, we then met at 12:30 for the Ukraine SIG Luncheon. The featured speaker was Phyllis Berenson who gave a fascinating slide show about her two recent trips to Ukraine, visiting her ancestral towns. Alex Dunai who was Phyllis’ highly recommended guide, was in attendance.
After lunch, at 2:00 pm, we all got together again for the Ukraine SIG annual meeting, where first Janette, and then each of the Directors reported on what has happened under their jurisdiction during the year and what is coming. Below is Lara Diamond, Projects Director for Ukraine SIG, telling us how to become a project coordinator.
Some of my notes from these Ukraine sessions were:
- Do a town search to identify your towns.
- If no town coordinator, then you should volunteer. It’s not much work.
- Projects need to be funded, translated and indexed.
- Most records are in Russian.
- The Ukraine archives do not allow posting of documents
- Only way to see original documents is to be a coordinator or translator
- Once documents are indexed, the originals will no longer be available
- If you find one you want, you will have to purchase it from the archives.
There was a session unfortunately scheduled at the same time as the Ukraine SIG meeting, about Volynia, a province in the Ukraine. Fortunately, the Conference coordinators were informed about this and the session was changed to 3:30 pm. I attended and it was a presentation by Alex Denysenko, a researcher from Volynia who could locate records in the archives for you. He says most cemeteries in Volynia are in terrible condition and had examples in his slides. It does sound like there might be Mezhirichi documents, so that’s encouraging for me.
To top off the day, 12 members of our Jewish Ancestry in Volhynia District Facebook Group met at 6:45 for a Dinner together at the Garden Grove Restaurant in our Swan Hotel. Thanks go to Gary Gershfield for arranging this. We all had a wonderful time. To my surprise and delight, I did not realize that this resort or this restaurant had character dining. I did not hesitate to get the necessary pics.
I also saw Bruce and Laurie Buzbee at the Garden Grove and got to talk to them again.
Wednesday
At 8:15 was the Computer Workshop: Getting Started with GEDmatch. The person originally scheduled to give the workshop, Rebecca Canada, ended up being unable to make it to the conference. So Sandy Aaronson was asked to lead the workshop in her place. Sandy, who assisted me during my workshop, asked if I would do so during hers, and of course I accepted. It was also a sneaky way for me to get into a GEDMatch workshop which I hadn’t signed up for.
The workshop ended at 9:45. I then had plans to join my younger daughter, who was working for the summer at Disney World through their Disney College Program. We went to Epcot and had lunch together at the Coral Reef Restaurant in the Living Seas attraction.

I got back in time to join the Romania SIG meeting at 5:00 p.m. Daniel Horowitz of MyHeritage led the meeting. I didn’t realize Daniel had any Romanian in him, but in fact he shares an ancestral place with me (Dorohoi), so maybe we’ll one day find that we’re related. The SIG leader, Barbara Hershey couldn’t make it … physically that is. She attended virtually on Daniel’s laptop.
There were about 30 in attendance, which was the largest ROM-SIG meeting yet. We talked about records, projects, towns. Jay Sage volunteered to take on the job of ROM-SIG webmaster, but he’s also the new Vice President of the IAJGS, so he’ll be busy.
At 8 p.m. was yet another event that I was really looking forward to. The film “Aida’s Secrets”, sponsored by MyHeritage, was about two brothers separated at birth and given up for adoption. It was through genealogical research by Yad Vashem and MyHeritage that they found each other. What was of great interest to me was that one of the brothers, Shepsel Shell was well-known in my Jewish community of Winnipeg in Canada, went to my synagogue when I was growing up, and lived next door to my brother-in-law when he was growing up. Many scenes from the film were taken in Winnipeg.
Here’s an article from the Winnipeg Free Press from 2016 about this: “A secret revealed – Shepsel Shell’s family reunification subject of a new doc”.
But it was the movie itself and the their mother’s “secrets” that were even more intriguing. Nothing is entirely clear. Following the movie, Daniel led an hour of boisterous discussion. Everyone had a different idea what might have happened.
In attendance to answer questions was Sima Velkovich from Yad Vashem in Israel who did all the Bergen-Belsen research. Also attending virtually (on the big screen after the movie) was British genealogist Laurence Harris whose research helped reunite the brothers.
Even though I had not seen the movie yet, my brother-in-law’s mother had, and she sent me to this conference with a number of questions to raise.
Neither Daniel nor the two experts could answer all the questions posed.
I’ll have lots to discuss with my brother-in-law and his mother when I see them this Friday.
Thursday
I started Thursday morning on a fun note. I met Ron Arons for the first time a few days earlier and he and I just hit it off – we’re both the same type A people. When he showed up in that penguin costume on Sunday night, it just cracked me up. So I couldn’t pass up the opportunity to hear Ron speak at 8:15 on “Finding Living People on the Internet”. I got there a few minutes late, and wanted to take a picture of him talking. He pointed me out and sternly scolded me and said, no photography. But in the end, he gave me full permission to post this picture of him speaking:
p.s. You can’t hide, because Ron will find you.
Following Ron’s talk, I went to the Professional Genealogist’s get together. There were 27 of us there. Most were APG members. Jeanette Rosenberg from England led the discussion. I knew many of the attendees, but it was especially nice to meet and talk to Gary Mokotoff and Stanley Diamond in person.
I then had a few hours in the Resource Room. Stationed in the room was Sima Velkovich from Yad Vashem who was at the Aida’s Secret movie the night before. I had a nice conversation with Sima, and she even was able to provide me with some Bergen-Belsen documents relevant to the movie that I can take home and review. Maybe I’ll get some ideas to answer some of the unanswered questions.
I tried a few of the subscription databases that were available for free in the Resource Room that I hadn’t tried before. Didn’t come up with anything significant. A few of my queries led me to my own postings such as my Family Research web page.
Then I took a few hours off to do visit a few Disney sites. I took a Disney bus to Animal Kingdom so that I could transfer to Coronado Springs, a Disney resort I was at 18 years ago with my family. From there to a bus to Epcot, where I boarded the monorail and took a couple of loops. I got off at Magic Kingdom and would have taken a bus to Hollywood Studios (to make it 4 parks in 4 hours), but I had run out of time and just headed back to the Swan to make it back in time for the banquet.
There was a pre-banquest reception starting at 6:30 in the Swan Foyer. Then at 7:00 pm, Adam Brown opened the Gala banquet:
The featured speaker was Professor Henry Louis Gates, Jr., host of Finding Your Roots. He spoke prior to the meal, possibly for an hour. He talked on genealogy and genetics in America. What an eloquent, intelligent, entertaining speaker. It was a pleasure to listen to him. Apparently, it was the first time he ever spoke at a genealogy conference. And he said afterward that he really enjoyed the evening. His series airs again this fall on PBS starting October 3. Don’t miss it.
Friday
The conference still had a half-day of programming left. Personally, I hate the end of a conference. I find it so sad. So I like to leave a little early, and knowing that the banquet would be the highlight of the week, I set up my flight home for Friday morning.
But I had one last treat. Janet Akaha was on the same shuttle bus to the airport with me. It was nice to have a good conversations with her and for me to learn a little more about Y-DNA in the process.
That’s that. A very full, fun, and educational week. Next up is Halifax in October.

































 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





