Genetic Clusters and DNAGedcom - Sun, 20 Jan 2019
Over the past 6 months, everyone has been jumping on board using genetic clustering techniques to help them partition their DNA matches into their ancestral origins.
The basic idea is to compare all the people that each of your DNA matches also match to. These are not segment matches being compared, but are people who are considered to be DNA matches to each of your DNA matches. These are known as the DNA testers who are In Common With (ICW) someone, i.e. they match each other.
The Leeds Method
The genetic clustering revolution started last summer with Dana Leeds who came out with the technique now named after her called The Leeds Method. It is primarily aimed at AncestryDNA testers, but will work with all companies. It really helps at AncestryDNA because they do not provide segment match information and have fewer tools that help you identify commonalities between your matches. The Leeds Method specifically is designed to partition your matches into groups representing each of your grandparents families.
Dana’s technique is a manual procedure and takes time to go one-by-one through your matches at AncestryDNA and add them to a spreadsheet. But it is a great exercise as it gives you a real feeling for how your matches relate to each other. It often works even if you have endogamy in your matches as I do.
When I tried it, I came out with this:
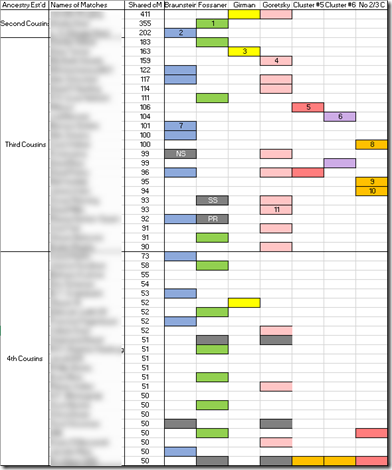
I was able to place the majority of the people in a Cluster (column). The first four columns I could identify as belonging to my four grandparents, which I show by their surnames in the top row. This technique can include relatives down to your closest 4th cousins, so I limited my matches to those who were 50 cM or more. The procedure takes several hours to do by hand. And you’ll probably, like me, do it incorrectly the first couple of times.
Some smart programmers were inspired by Dana’s method. Doing it by hand is laborious, so why not automate it, they thought? And while at it, why not figure out a great way to visualize it.
Genetic Affairs
In November, Evert-Jan Blom put his mind to this and developed Genetic Affairs. It is an online program that logs into your Ancestry account, gathers the ICW data, and produces your clusters in a large table that lists all your DNA matches on both the left and the top. If they match in a cluster, they are given a color for the cluster. If they are a match outside all clusters, they are colored grey.
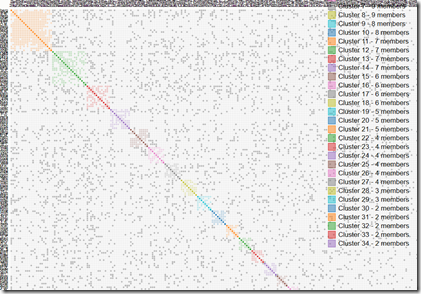
Your match table gets emailed to you when it is ready. Mine looked like this:
Although there are lots of grey squares (representing my endogamy), there are also 34 colored clusters. Using my results from the Leeds Method helped me identify the ancestors for several of the clusters and I was able to figure out where some of the others were from as well. I ended up being able to assign about 90 of my AncestryDNA relatives to a particular ancestor in 12 of the clusters.
DNAGedcom – First Download Attempt
In December, over at DNAGedcom, they added the Collins’ Leeds Method 3D. Kitty Cooper describes it nicely. To try it, I had to resubscribe to DNAGedcom for their $5 a month fee.
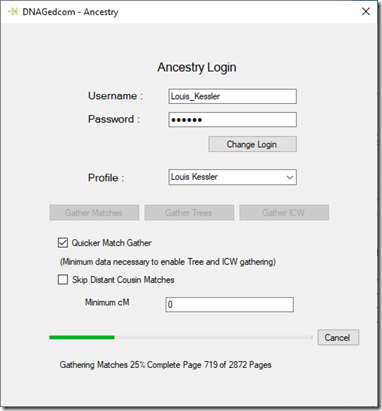
Then I had to download some data. You do this with the DNAGedcom Client running on your Windows computer. And then, if you have as many matches as I do, you wait patiently. Here is one of the progress windows:
I’ve got 2,872 pages of AncestryDNA matches to download, which equals about 143,600 people.
The defaults were “Quicker Match Gather” selected, “Skip Distant Cousin Matches” unselected, and 0 for “Minimum cM”. I left them all as their default. I thought that’s probably a mistake, but what the heck. My computer’s not doing much right now anyway.
There are three steps to download AncestryDNA data using DNAGedcom Client:
Step 1, Gather Matches. I started the “Gather Matches” step of the program at 9:54 a.m. I was able to do other work on the computer while it was running (i.e. work on Version 3.0 of Double Match Triangulator). The Gather Matches step finished at 12:49 p.m. and the resulting DNAGedcom.db (database file) is 80.6 MB. So in total, it took almost 3 hours. That worked at a speed of just over 16 pages of 50 people per minute. The database uses on average 589 bytes per person.
The Gather Matches step also created a 46.9 MB file called m_Louis_kessler.csv. This file contains a title line row plus 143,583 rows, one for each of my Ancestry matches. The columns are:
- testid – some unique long identifier representing me.
- matchid – some unique long identifier representing my match.
- name – the name of the tester I match to.
- admin – the name of the person who is administrator for the test. The name is the same as the admin for 120,536 people (83.9%) in my file.
- people – the number of people in their tree. Of my matches, 59,581 (41.5%) have trees. 42,406 (29.5%) have at least 10 people in their tree. 20,393 (14.2%) have at least 100 people in their tree. 28 trees have over 100,000 people in it. The largest has 277,652 people. The total number of people in my 59,581 matches’ trees is 42,595,797, averaging 715 per tree. You’d think there should be a good number of relatives in there for me to find.
- range - Relationship range, I have 28 second and third cousins, 13,769 fourth cousins, and 129,786 distant cousins.
- confidence – a number that’s 100 for my first 2 matches and goes down to 21.198 for my last match.
- shared cM – for me ranges from 410.8 down to 6.
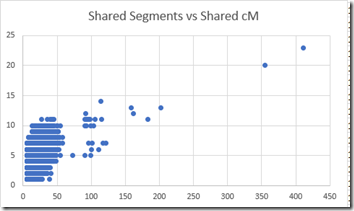
- shared segments – for me, from 23 down to 1. I have 19,191 sharing just one segment. Here’s an XY plot:
- last login – there is nothing in this column for me
- starred – if I’ve starred the person, true or false
- viewed – if I’ve viewed the person, true or false
- private – if the person is private. 9,084 (6.3%) of mine are marked private
- hint – not sure what this is, but all of mine are false
- archived – there is nothing in this column for me
- note – there is nothing in this column for me. I don’t use Ancestry notes.
- imageurl – a link to the DNA tester’s profile picture at Ancestry. I have profile pictures on 16,016 (11.2%) of my matches.
- profileurl – there is nothing in this column for me
- treeurl – a link to the DNA tester’s tree at Ancestry. I have trees linked from 60,090 (41.9%) of my matches.
- scanned – this column has today’s date for every match, since I started from scratch and got all my matches today. If I run DNAGedcom client again in the future, I can use this column to identify the new matches since my earlier run.
- membersince – there is nothing in this column for me
- ethnicregions – there is nothing in this column for me
- ethnictraceregions – there is nothing in this column for me
- matchurl – a link to the DNA tester’s match page at AncestryDNA.
I bet if I unclick the option “Quicker Match Gather”, that the columns that are now empty for me will get filled. I don’t need them for the clustering, so I’ll try that some other time.
Step 2, Gather Trees. I started the “Gather Trees” step of the program at 12:57 p.m.to gather 51,006 trees. This ran all afternoon and finally finished in the evening 11:42 p.m. That step took 10 hours and 45 minutes. That was an average of 79 trees per minute. The database size grew to 291.6 MB, which is an increase for this step of 211.0 MB. So the average tree needed about 5,994 bytes in the database.
Step 2 finished by creating a 162 MB file named a_Louis_Kessler.csv. This file has a title line followed by 1,028,007 rows of data. That’s pretty close to Excel’s limit of 1,048,576 rows. A few more trees, and I wouldn’t have been able to open that file with Excel but would have had to manually divide it into pieces with a text editor first and then load it in parts. The columns in this file are:
- testid – some unique long identifier representing me.
- matchid – some unique long identifier representing my match. There are only 50,780 different matchids in the file. This is a bit less than the 51,006 “trees” DNAGedcom said it was loading. I’m not sure why.
- name – the name of the tester I match to. Because there are 50,780 different testers in the file, the average tester has 20 lines. These cannot be the full trees of the people. If they were, I’d be looking at 715 people per tree (see “people” in the “Gathering Matches” section, above). So it seems obvious that these are just the ancestors of each person from their trees.
- admin – the name of the person who is administrator for the test
- surname – the surname of an ancestor of the tester. 875,392 (85.2%) of these had a surname in it. The rest were blank. The most common surnames for me were Cohen (6,408), Smith (4,072), Miller (2,919), Schwartz (2,698), Goldberg (2,632), Brown (2,628) and Levy (2,546), which is a good mix of what happen to be the most common Jewish and non-Jewish surnames. With regards to some of my own ancestors surnames, there’s Braunstein (145), Focsaner (0), Goretsky (6), Silverberg (115). I took a look at some of these and I cannot connect any of them to my ancestors. There are other spellings of these names as well. As far as Kessler goes, there’s 392, but that does not matter here because I’m not DNA related to Kessler, who was my father’s stepfather.
- given – the given name of the ancestor. 111,506 (10.8%) of these say “Private” and have a blank surname. Only 17,854 (1.7%) of the given names are blank. The most common given names for me were John (15,487), Mary (15,223), Sarah (14,220), Elizabeth (11,829), Samuel (11,348), Joseph (11,260) and William (11,239). I personally found it interesting that Louis was in 16th place with 5,783.
- birthdate – 704,463 (68.5%) have values.
- deathdate – 581,187 (56.6%) have values.
- birthplace – 679,101 (67.8%) have values. The most common are Russia (52,579), New York (12,887), Poland (11,403), Austria (9,439), Germany (8,676). All my ancestors come from either Romania (4316), specifically Tecuci (5), Dorohoi (23) or Ukraine (1,435), specifically Mezhirichi (3) or Odessa (736).
- deathplace – 558,889 (54.3%) have values.
- relid – this looks like it is the ancestor’s ahnentafel number, which is 1 for the tester, 2 for the tester’s father, 3 for the mother, 4 for their paternal grandfather, 5, for paternal grandmother, etc. The highest number is 1023 which is the person’s mother’s mother’s … mother (with 9 mothers – i.e. 7th great-grandmother, 9th generation). For the 50,780 people, all of them list themselves. So that must be the reason for the cutdown from 51,006. Those 226 people must not have had themself in their tree. The average parent is included 47,528 times (93.6%), grandparent 40,070 (78.9%), 3rd Gen: 22,364 (44.0%), 4th Gen: 9,736 (19.2%), 5th Gen: 3,374 (6.6%), 6th Gen: 1,266 (2.5%), 7th Gen: 542 (1.1%), 8th Gen: 254 (0.5%), 9th Gen: 124 (0.2%)
- source – there is nothing in this column for me.
Step 3: Gather ICW. ICW stands for the “In Common With” people. This is what is used to cluster all the people using the various clustering techniques. I started this procedure at 11:42 p.m. The screen indicated that it was going to find the ICW for all 143,583 people. It wasn’t progressing very quickly. By 12:10 a.m, it had only completed 98. So I went to bed. When I checked in the morning at 8:19 a.m., DNAGedcom had completed only 1,393 (0.9%) of the ICWs. It was averaging 160 people per hour. The database had grown from 291.6 MB to 854.5 MB, and increase of 562.9 MB which is 5.7 MB per person. If it continued at this speed, it was going to take 37 days and nights for it to complete, and the database would become 819 GB in size. Now yes, maybe as it goes to the more distant relatives, it might speed up and contribute less to the database. So I thought I’d see if it would. DNAGedcom even as it was running, allowed me check the Skip Distant Cousin Matches. So I thought maybe it would recognize that and stop after my 13,797 second, third and fourth cousins. I then let it continue run for the day while I was out of the house. When I checked it at 6:02 p.m., it had only done 2,891 (2%). It was not going any faster and was still averaging about 160 people per hour. It still said its goal was 143,583 people. I didn’t want to let it run another 3 days to see if it would stop at 13,797, so I disappointingly hit the cancel button. And then I was again disappointed when I saw that no file was generated. I knew it was supposed to create an ICW file when it completed this step. That is the file that is used for the clustering procedures. I was hoping DNAGedcom would still generate this file with what it already had processed up to cancelling, but it didn’t.
DNAGedcom – Second Try
I saved my old files and this time from the beginning, selected “Skip Distant Cousins” and also set Minimum cM to 20, which is the level AncestryDNA starts its Distant Cousins at. I then thought I might as well uncheck the “Quicker Match Gather” and see what additional information might be retrieved.
Step 1: Gather Matches. Started at 6:10 p.m. It processed 2,880 Ancestry pages and finished by 8:04 p.m. So that was an hour quicker than previously. The database was 13.0 MB, one fifth the size it was previously. What I found extremely interesting was that it processed 2,880 pages. Just a day ago, it processed only 2,872 pages. Those 8 pages represent about 400 more matches that I have gained in just one day! AncestryDNA must have sold a lot of tests during the holiday period and the results are starting to come in!
The Gather Matches file m_Louis_kessler.csv is now just 6.2 MB in size. It now lists just my 13,843 fourth cousins and closer. I had 13,797 just a day earlier. With the “Quicker Match Gather” turned off, I now have information in these columns:
- profileurl – this now has values
- membersince – lists the year the tester became a member of Ancestry. 28.7% were between 2010 and 2015. 11.7% were 2016. 22.0% were 2017. 22.3% were 2018. 15.4% were 2009 and earlier with the earliest year being 2000 by 110 people (except for 80 people listed obviously wrongly shown as becoming a member in the year 1900)
- ethnicregions – these are lists of the top ethnicities of each match. For me, the number one ethnicity of my matches is “EuropeJe”, i.e. European Jewish and 10,707 (74.4%) are listed as that. Another 2975 (21.5%) have “EuropeJe” followed by one or more other ethnicities (e.g. Slavic, EuropeS, Baltic, Germany, etc.), and 156 (1.1%) have “EuropeJe” in their list but not listed first. Only 1 match listed as: “Celtic, EuropeW, EuropeE, EuropeS, AngloSaxon” does not have “EuropeJe” in their list of ethnic regions.
- ethnictraceregions – this had values for 8804 (63.6%) of my matches are were all over the map with no discernable trends.
Step 2: Gather Trees: I started this at 8:13 p.m. It finished gathering 4,819 trees at 9:15 p.m. That was an average of 78 trees per minute, which was the same speed as previously, only there were fewer trees to process this time round. The database grew by 21.1 MB to 27.3 MB.
This time Step 2 created a_Louis_Kessler.csv as a 10.9 MB file with 72,491 lines for 4,802 people.
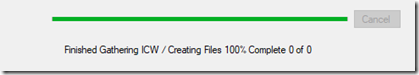
Step 3: Gather ICW. I started this at 9:21 p.m. Two hours later, at 11:17 p.m., it was only at 302 of 13843, just 2.1%, I was calculating that it would take 98 more hours. I was seriously considering stopping it, and upping the limit on Minimum cM and trying this for a third time. But then, at 11:22 p.m., the DNAGedcom progress indicator changed to 100% saying: “Finished Gathering ICW / creating FIles 100% Complete 0 of 0”
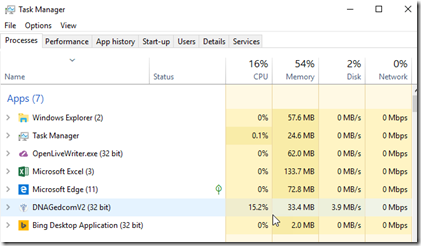
What was strange here was the statement: “0 of 0”. It was supposed to create an ICW file that I can use. There was no such file created.It looked like DNAGedcom had finished. But before I gave up hope, I opened Task Manager:
Task Manager showed DNAGedcom was still using CPU and writing to disk. Maybe it was still creating that ICW file, but just not telling me that it was.
Sure enough, at 11:45 p.m, it completed and created the icw_Louis_Kessler.csv file. That took 23 minutes. Finally DNAGedcom displayed:
DNAGedcom’s progress indicator is misleading. When you Skip Distant Cousin Matches or set a Minimum cM, it should show you progress relative to that, and not suddenly change you from 2.1% to Completed. It should then tell you that it is creating the ICW file. The statement: “Creating Files 100% Complete 0 of 0” is not an indicator that there is still something being created, especially when the phrase “100% Complete” is in the middle of it.
None-the-less, I now have my ICW file. It is 119.2 MB and contains 924,299 lines. The columns are:
- matchid – the unique long identifier representing this match
- matchname – the name of the tester of this match
- matchadmin – the person administrating this match
- icwid – the unique long identifier representing the ICW match
- icwname – the ICW’s name
- icwadmin – the ICW’s admin
- Source – the value for all lines is “Ancestry”. DNAGedcom works with other company’s data as well, thus the reason for this column.
My file contains 13,767 different matches who are ICW on average 67 other matches each. The most any match is ICW is 3,749 (27.2% of my 4th cousins and closer). I have 240 matches ICW 1,000 or more people. I have 93 matches ICW with only 1 other match, and 128 matches ICW only 2 other matches.
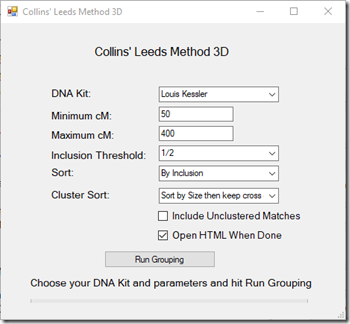
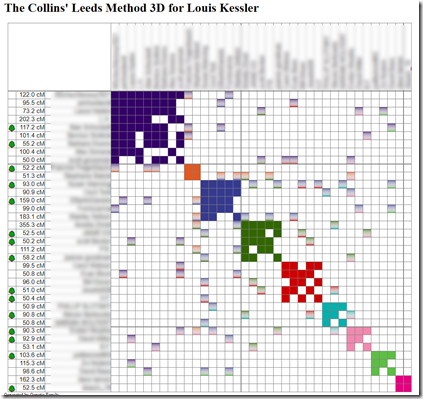
Collins’ Leeds Method 3D
It will now be interesting to see what DNAGedcom’s new clustering algorithm does with my ICW information.
It gives me this, which very much corresponds to the manual Leeds method I first used:
In order to attempt to do something similar to what Genetic Affairs does, I lowered the Minimum cM to 40 and got this:
So yay! Clusters are real and are useable to partition your DNA relatives into groups, even if you come from endogamy like I do.
Shared Clustering
Just this month, another clustering approach was developed using Ancestry data from DNAGedcom downloads. This is an open source program called Shared Clustering built by Jonathan Brecher and is available at Github.
On his ”Shared Clustering versus other clustering tools” page, Jonathan states:
As of this writing, the clusters generated by Shared Clustering are significantly better than those generated by most other tools. In this context, "better" means that the clusters are more useful to the genealogical researcher.
and he follows by describing the reasons in detail. Obviously, this is something that needs to be tried.
I downloaded and ran the setup.exe program from the Shared Clustering Github site. Windows gives you a warning because the author did not code sign the program, but by pressing “more info” on the warning, I could then click “run anyway". After ignoring two more such warnings, it installed.
The main screen starts with an Introduction:
Since I had my ICW file from DNAGedcom, I went to the Cluster page. I put the path to my ICW file in the Saved data file box, and it automatically entered a cluster output file the same directory with the name: Louis_Kessler-clusters.xlsx
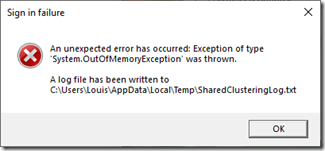
It ran very quickly, but before it finished, it gave this error:
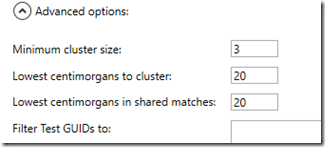
Obviously, it couldn’t handle the size of my ICW file. So I went to the advanced options:
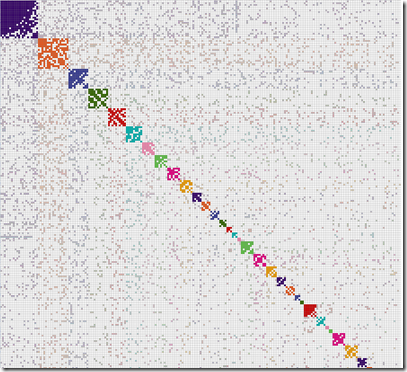
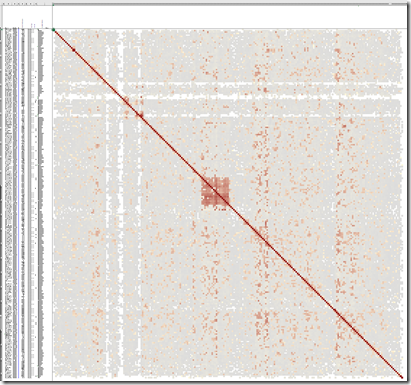
I changed both the 20 values to 40 and ran it again. This time it worked. I went the directory with the output files and opened up the xlsx file. Here’s what it looked like at 15% magnification:
There are 243 people included in 7 clusters. I can’t offhand identify my ancestors for that dark cluster 4 in the middle.
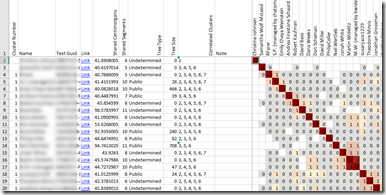
Interestingly, it gives extra information at the beginning of each row:
This includes tree information which is not in my ICW file, so it must be reading one of the other DNAGedcom files as well (or the DNAGedcom database). It also includes a list of correlated clusters for each person.
Conclusion
I’ve compared the visual results from:
- The Leeds Method
- Genetic Affairs
- Collins’ Leeds Method 3D
- Shared Clustering
Each gives useful results that can help you cluster your AncestryDNA matches into possible groups that have a common ancestor. Tools to help you at AncestryDNA are important because Ancestry does not give access to your segment matches or a chromosome browser. So clustering can help you determine possible ancestral lines that groups of DNA relatives may share with you, and that will help you direct your genealogical research to connect yourself with them.
These ideas for using In Common With (ICW) data translate well to Chromosome Mapping and Triangulation techniques. Double Match Triangulator already shows you ICW data on the People page for all the B People in a combined run, added who among them triangulates with each other. More uses of ICW data are going to be in DMT 3.0 as I work to finish and release it.

 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





