As I noted in my last post, Whole Genome: The VCF File, Part 2, the SNP VCF (Variant Call Format) file that Dante Labs gives tester of WGS (Whole Genome Sequencing), does not quite have everything that is needed to generate a raw data file that can be uploaded to various DNA sites for people matching.
The VCF file contains SNPs that vary from the standard human reference genome. The thinking is then that any SNPs needed for the raw data file (to match with the SNPs that are tested by the chips used by Ancestry, 23andMe, Family Tree DNA, MyHeritage DNA and Living DNA) that are not in the file can simply be replaced by the value from the standard human reference.
But they can’t.
The problem is that the VCF contains only those variants that passes a set of filters meeting some quality controls to ensure that the read was good enough. Those SNPs that fail the filters are not included in the VCF file. So some actual variants that should be in the VCF file don’t make it. These are known as true negatives. Substituting the standard reference genome values for those will give you incorrect values.
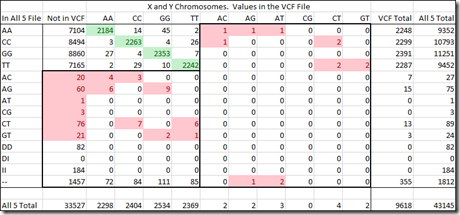
How many might there be? Well take a look again at the table from my last post.
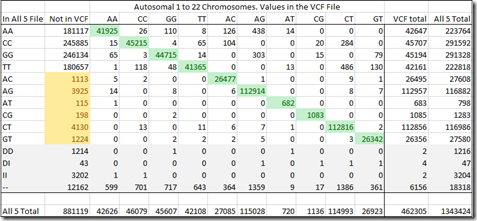
When I created my “All 5” raw data file, it ended up containing 1,343,424 SNPs.
My VCF file contains 3,442,712 variant SNPs. Of those, 462,305 (13.4%) were at positions in my All 5 file.
The green highlighted values are those that matched the consensus of my All 5 file. If you exclude the no-calls and deletions in my All 5 file, then that’s 453,534 out of (462,305 – 2 – 4 – 2 – 6,156 =) 456,141 or 99.4% agreement on the values. Only 0.6% disagree which isn’t too bad.
But the real problem are those yellowed values. Those are SNPs with two different allele values (heterozygous) and they by definition must be variants. Yet, the VCF file does not include them. These amount to another 10,705 SNPs that either were wrong in the chip reads, or should have been in the VCF file but were not. This means that as many as 2.3% of the VCF values may not be included in the VCF.
There could be others as well. The AA, CC, GG and TT values just above the yellow cells that are not in the VCF may have been variants but not included in the VCF file. e.g. If the reference value was G and the SNP value was AA, then this homozygous read is a variant and should be among the green values in the AA row and AA column. But the VCF may not contain it. We can’t tell how many of these there might be until/unless we get a third test done to compare.
It’s for these true-negative values, that substituting the reference genome value would be an incorrect thing to do and build you a substandard raw data file with likely thousands of incorrect values in it.
The Raw VCF File
Dante gives you access to your filtered VCF file. There is another file available that you can get. If you copy the link to your Dante Labs VCF file and change the end of the filename from “snp.vcf.gz” to “raw.snp.vcf.gz”, you can download what I’m calling the Raw VCF file.
This file contains the unfiltered variants found by your whole genome test. None of the quality measures were applied to this file. The quality filtering that Dante does is designed to get remove most of the false positives, i.e. to prevent SNPs that are not variants from being reported to be variants.
My Raw VCF contains 3,973,659 SNPs. It includes every one of the 3,442,712 SNPs in my filtered SNP VCF file, plus the 712,064 SNPs (17.9%) that it found that it filtered out because the quality didn’t wasn’t enough to be sure they were true variants.
So you can use this Raw VCF file to get more of your true negatives back, but by doing so, you will also add false positives to it.
It’s a tricky situation. It’s a trade-off like a teeter totter, with false positives on one side and true negatives on the other. Dante picked a set a filters that presumably take a good compromise position that does its best to minimize the two types of mistakes.
The one nice thing about this Raw VCF file is that it includes my 46 mtDNA variants. The VCF filters somehow removed these and they are not in the filtered VCF file. Once I got these, I was able to put them into 23andMe raw data format and upload it to James Lick’s mtDNA Haplogroup utility. The format I used is:
i703360 MT 114 T
Since the raw file doesn’t give the RSID, I just used i703360 for everything, hoping that James Lick didn’t use it, and it appears he doesn’t. I used MT as the 2nd field because that’s what 23andMe has. 114 is the position number. T is the value. Those are Tabs (Hex 09) between the fields, not spaces.
I know these were my mtDNA variants because Lick’s routine gave me my correct mt haplogroup: K1a1b1a.
However, for most purposes, this Raw VCF File is likely worse to use than the filtered VCF file since it will include more false positives (variants that are not really variants) than will the filtered VCF file.
And the Raw VCF File also won’t help in our goal to produce a raw data file that can be uploaded to sites for people matching. Reducing true negatives is good, but increasing false positives is bad.
The Genome VCF (gVCF) File
So we still want/need/hope for a way to produce that raw data file from our WGS results. Too many errors are introduced by adding human reference values where they don’t have values in the VCF file. Using the Raw VCF file will alleviate some of the true negatives but it also will increase the false positives, which is not a good trade-off.
There is an intermediate file. It is called the gVCF or Genome VCF file. It contains all the reads of the Raw VCF file and then is said to fill in the gaps with the human reference genome.
Well, that really doesn’t help. It is still basically the Raw VCF file. All it does is supposedly make the lookup of the human reference genome a little simpler.
I requested my gVCF from Dante and they made it available to me. It was 3.6 GB in size and took 30 minutes for me to download. It is a compressed file and took 5 minutes to unzip. The full file is 24.6 GB
Here’s a snippet from it:
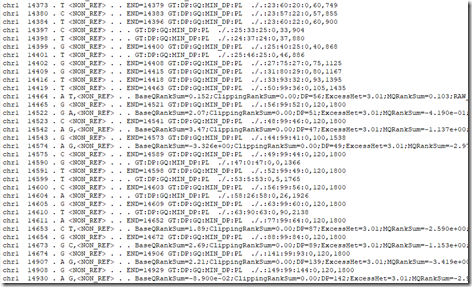
The long lines are the variants. The short lines are the positions where I don’t have a variant.
Position 14464 on Chr 1 is the first variant in my filtered VCF file. This file contains all the variants in my filtered VCF file.
Positions 14464 and 14653 are my 6th and 7th variants in my Raw VCF file. This file contains all the variants in my Raw VCF file.
But positions 14523, 14542, 14574, 14673, 14907 and 14930 are variants that are in this file, but not in either my Raw VCF file or my filtered VCF file. So there must even be more filtering done even before the variants even make it to the Raw VCF file. Maybe some of the values on the line (like the BaseQRankSum for example) indicate uncertain reads and have something to do with them being left out. None-the-less, I wouldn’t get excited about adding these additional variants because many likely will be false positives if you do.
At least the hope is that you’ll get every read in every position from this file.
But no. It doesn’t even do that.
The lines given that are not variants often contain an END value. e.g., the first 3 lines I displayed above contain:
14373 . T END=14379
14380 . C END=14383
14384 . T END=14396
Does the value T represent all the values from positions 14373 to 14379, or does it just represent the first position? My perusal of the VCF specs finds:
The value in the POS field refers to the position of the first base in the String.
To verify, I took some homozygous SNPs from my DNA tests that agree between different companies:
Chr 1: position 1040026 was read as TT by all 5 companies. My gVCF has:

So it says C at position 1039779, ending at position 1040048. That’s not T.
Try another: Chr 1: position 1110019 was read as AA by all 5. My gVCF has:

Here it says C at position 1109397, ending at position 1110155. That’s not A.
So the value shown refers to the first position. You do not know what the other positions up to the end positions hold.
And in fact, my gVCF file contains 308,122,966 data lines. That is about 10% of the 3 billion base pairs we have. So only 1 out of 10 positions are reported in the gVCF file with either your variant value or the human genome value.
None-the-less, it doesn’t matter whether the gVCF file contains all the human reference genome values or not. The variants in it are even more liberal than the Raw VCF file and would introduce even more false positives if it is used the generate a raw data file.
A Possible Solution
Between the filtered VCF, the Raw VCF and the gVCF files, none of them alone have the data in it to generate a raw data file that can be uploaded to GEDmatch and other DNA testing sites. And they have a sliding range of variants from optimistic (gVCF) to liberal (Raw VCF) to conservative (filtered VCF).
The problem is that DNA results include both false positives and true negatives. DNA testing companies get around the uncertain values by indicating them to be no-calls. That works because no-calls are always considered a match, and therefore they won’t break a matching segment. As long as there are not too many of them (i.e. 5% or less), the segment matching should work well.
So I believe we can generate a raw data file by doing this:
- Make a list of all the SNPs you want raw data for.
- Initially assign them all the human genome reference values. Note: none of the VCF files give all of these to you, so you need this initially set this up. Wilhelm HO have a good set of them included with his DNA Kit Studio.
- The positions of variants in your gVCF file should be marked as no-calls. Many of these variants are false, but we don’t want them to break a match.
- The positions of variants in your filtered VCF should be marked as having that variant. This will overwrite most of the optimistic no-calls marked in step 3 with filtered reliable values.
I likely will try this myself. When I do, it will be worthy of its own blog post.



 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





