Finally, Interesting Possibilities to Sync Your Data - Fri, 17 May 2019
Although I don’t use Family Tree Maker (FTM), per se, I am very interested in its capabilities and syncing abilities. FTM along with RootsMagic are the only two programs that Ancestry have allowed to use the API that gives them access to the Ancestry.com online family trees. Therefore they are the only two programs that can directly download data from, upload data to, and sync between your family tree files on your computer and up at Ancestry.
RootsMagic
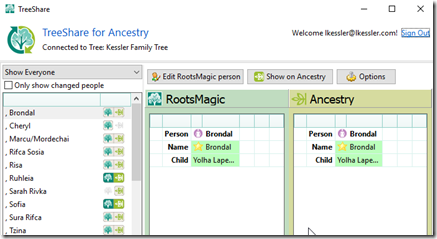
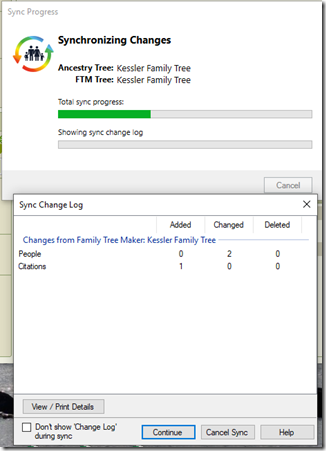
RootsMagic currently has its TreeShare function to share the data between what you have in RootsMagic on your computer, and what you have on Ancestry. It will compare for you and show you what’s different. But it will not sync them for you. You’ll have to do that manually in RootsMagic, one person at a time using the differences.
That is likely because RootsMagic doesn’t know which data is the data you’ve most recently updated and wants you to verify any changes either way. That is a good idea, but if you are only making changes on RootsMagic, you’ll want everything uploaded and synced to Ancestry. If you are only making changes on Ancestry, you’ll want everything downloaded and synced to RootsMagic.
With regards to FamilySearch, RootsMagic does a very similar thing. So basically, you can match your RootsMagic records to Family Search and sync them one at a time, and then do the same with Ancestry. But you can’t do all at once or sync Ancestry and FamilySearch with each other.
With regards to MyHeritage, RootsMagic only incorporates their hints, and not their actual tree data.
Family Tree Maker
Family Tree Maker takes the sync with Ancestry a bit further than RootsMagic, offering full sync capabilities up and down.
For FamilySearch, FTM up to now only incorporates their hints and allows merging of Family Search data into your FTM data, again one person at a time. But Family Tree Maker has just announced their latest upgrade, and they include some new FamilySearch functionality.
What looks very interesting among their upcoming features that I’ll want to try is their “download a branch from the FamilySearch Family Tree”. This seems to be an ability to bring in new people, many at a time, from FamilySearch into your tree.
Family Tree Builder
MyHeritage’s free Family Tree Builder download already has full syncing with MyHeritage’s online family trees.
They do not have any integration with their own Geni one-world tree, which is too bad.
But in March, MyHeritage announced a new FamilySearch Tree Sync (beta) which allows FamilySearch users to synchronize their family trees with MyHeritage. Unfortunately, I was not allowed to join the beta and test it out as currently only members of the Church of Jesus Christ of Latter-Day Saints are allowed. Hopefully they’ll remove that restriction in the future, or at least when the beta is completed.
Slowly … Too Slowly
So you can see that progress is being made. We have three different software programs and three different online sites that are slowly adding some syncing capabilities. Unfortunately they are not doing it the same way and working with your data on the 6 offline and online platforms is different under each system.
The very promising Ancestor Sync program was one of the entrants in the RootsTech 2012 Developer Challenge along with Behold. I thought Ancestor Sync should have won the competition. Dovy Paukstys, the mastermind behind the program had great ideas for it. It was going to be the program that would sync all your data with whatever desktop program you used and all your online data at Ancestry, FamilySearch, MyHeritage, Geni and wherever else. And it would do it with very simple functionality. Wow.
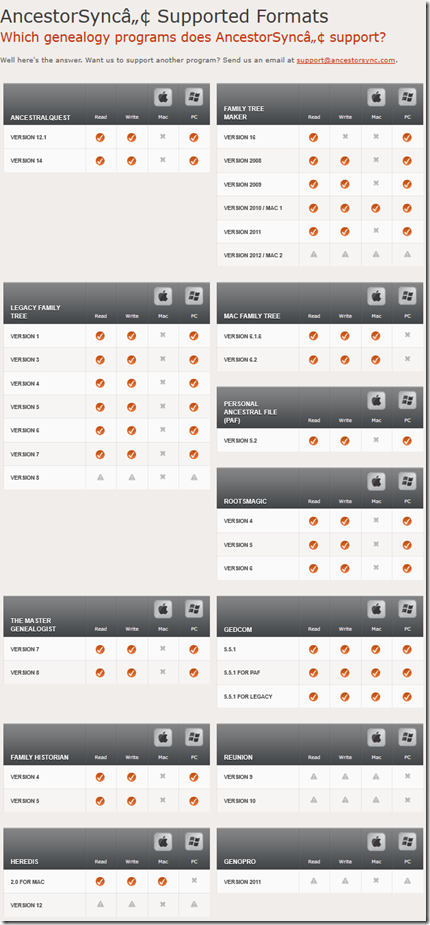
This was the AncestorSync website front page in 2013 retrieved from archive.org. 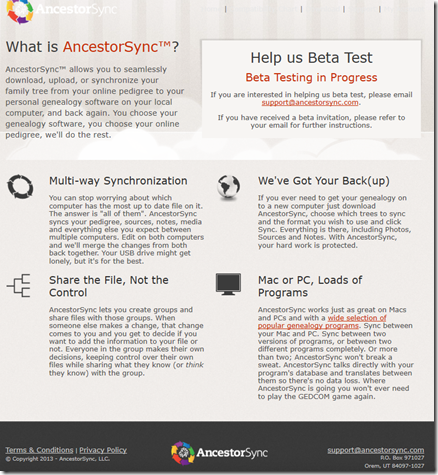
They had made quite a bit of progress. Here is what they were supporting by 2013 (checkmarks) and what they were planning to implement (triangles):
Be sure to read Tamura Jones’ article from 2012 about AncestorSync Connect which detailed a lot of the things that Ancestor Sync was trying to do.
Then read Tamura’s 2017 article that tells what happened to AncestorSync and describes the short-lived attempt of Heirlooms Origins to create what they called the Universal Genealogy Transfer Tool.
So What’s Needed?
I know what I want to see. I want my genealogy software on my computer to be able to download the information from the online sites or other programs into it, show the information side by side, and allow me to select what I want in my data and what information from the other trees I want to ignore. Then it should be able to upload my data the way I want it back to the online sites, overwriting the data there with my (understood to be) correct data. Then I can periodically re-download the online data to get new information that was added online, remembering the data from online that I wanted to ignore, and I can do this “select what I want” again.
I would think it might look something like this:
where the items from each source (Ancestry, MyHeritage, FamilySearch and other trees or GEDCOMs that you load in) would be a different color until you accept them into your tree or mark them to ignore in the future.
By having all your data from all the various trees together, you’ll easily be able to see what is the same, what conflicts, what new sources are brought in to look at, and can make decisions based on all the source you have as to what is correct and what is not.
Hmm. That above example looks remarkably similar to Behold’s report.
I think we’ll get there. Not right away, but eventually the genealogical world will realize how fragmented our data has become, and will ultimately decide that they need to see it all their data from all sites together.

 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





