Someone on Facebook reported a new feature at 23andMe and I couldn’t wait to try it. This 23andMe beta “auto-builds” your family tree from your DNA connections with other customers.
They aren’t the first to try something like this. Ancestry DNA has ThruLines, which uses your tree and your DNA match’s tree to try to show you how you connect. MyHeritage DNA does the same with their Theory of Family Relativity (TOFR) using your tree and your match’s tree at MyHeritage.
I had good success at Ancestry who gave me 6 ThruLines joining me correctly to 3 relatives I previously knew the connection to, and 3 others who were correctly connected but were new to me. I then contacted the latter 3 and we shared information and I was able to add them and their immediate relatives to my tree.
I had no success yet at MyHeritage DNA even though I have my main tree there. I’ve never had a sing TOFR there fore either me or my uncle. My closest match (other than my uncle) is 141 cM. My uncle’s closest is 177 cM. Those should be close enough to figure out the connection. But none of the names of my matches at MyHeritage give me any clues, and I haven’t been able to figure out any connection with any them, even using some of their extensive trees.
23andMe does not have trees to work with like Ancestry and MyHeritage. Actually, I shouldn’t say that. Not too long ago, in another beta, 23andMe allowed uploading your FamilySearch tree to 23andMe. See Kitty Cooper’s blog post about it for details about it. They never said what they were going to do with that data, but I wanted to be ready if they did do something, All it tells me right now is this:
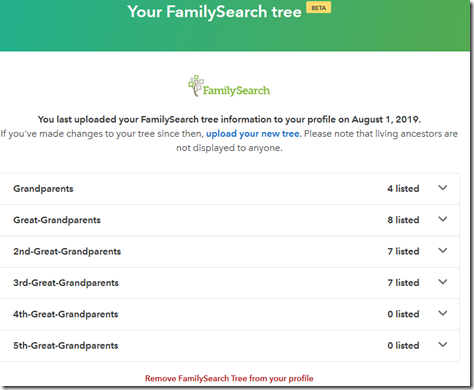
If you include your FamilySearch Tree in your profile, then anyone else who has done the same will show a FamilySearch icon next to their name. You can also filter for those who have done so. I don’t think many people know about this beta feature yet, because my filter says I have no matches who have done so.
But I digress. Let’s go check out the new Family Tree beta at 23andMe. I’m somewhat excited because I have a dozen relatives who I know my connection to who have tested at 23andMe. I’ve been working with them over the past 2 weeks getting my 23andMe matches to work in my (almost-ready) version 3.0 of my Double Match Triangulator program. And the odd thing about all my known relatives at 23andMe is that they are all on my father’s side! I’d love to be able to connect to a few people at 23andMe on my mother’s side. Maybe this Family Tree beta will help. Let’s see.
So I go over to my 23andMe “Your Family Tree Beta” page. It takes it a few minutes to build my tree and update my predicted relationships. Once it does, out pops this wonderful diagram for me. (Click on the image to enlarge it).
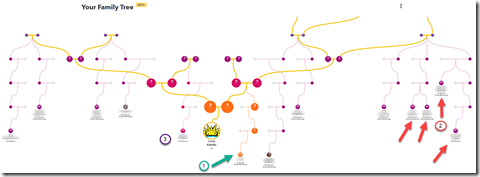
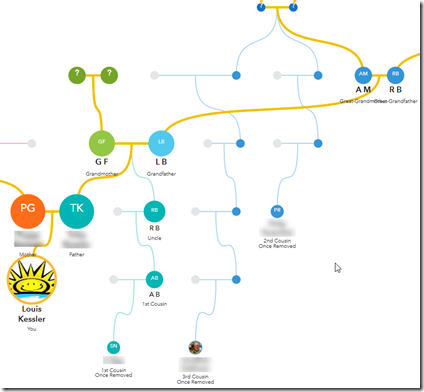
I’m shown in the middle (my Behold logo sun), with my parents, grandparent, great-grandparents above. And 23andMe has then drawn down the expected paths to 13 of my DNA matches.
This is sort of like ThruLines and TOFR, but instead of showing just the individual connections with each of the relative, 23andMe are showing all of them on just one diagram. I like it!!
The 13 DNA matches they show on the diagram include 5 of the 10 matches I have that I know my relationship with (arrows point to them), and 8 who I don’t know my relationship with. Maybe this will help me figure out the other 8.
My 3 closest 23andMe matches are included, who I have numbered 1, 2 and 3. Number 3 is on my mother’s side, but I don’t know what the connection is. None of the other 10 are on my first page of matches (top 25).
The number 1 with a green arrow is my 1st cousin once removed. She is the granddaughter of my father’s brother. So that means the entire right side of the tree should be my father’s side and the left should be my mother’s side.
The ancestors are all shown with question marks. I can now try labelling the people I know because of my connection with my first cousin. When I click on the question mark that should be my father, I get the following dialog box:
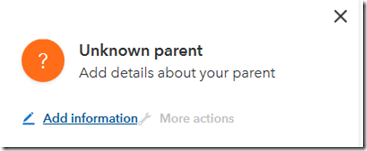
The “More actions” brings up a box to add a relative, but that action and likely others that are coming are not available yet.
When you click on “Add Information” you get:
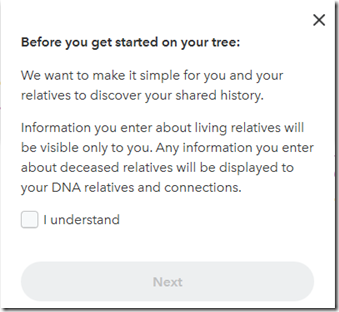
I click on “I understand” and “Next” and I get:
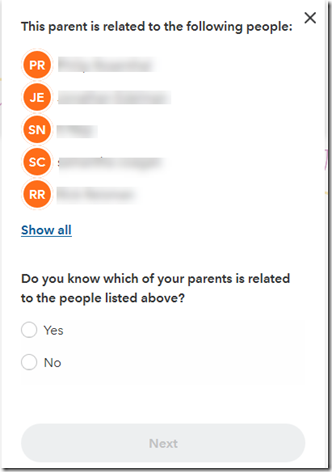
I click “Yes” and “Next” and it lets me enter information about this person:
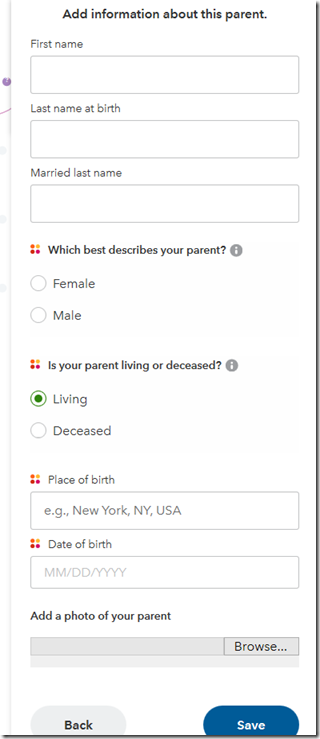
Now I’m thinking at this point that they have my FamilySearch info. Maybe in a future version, they can allow me to connect this person to that FamilySearch tree, and not only could they transfer the info, but they should be able to automatically include the spouse as well.
But for now, I simply enter my father’s information and press “Save”. I did not attempt to add a photo.
When I clicked “Deceased”, it added Place of death and Date of death. But it has a bug because the Place of Death example cannot be edited. But what the heck. This is a beta. Expect a few bugs.
It gives a nice confirmation box and then on the chart changes the orange circle with the question mark to the green circle with the “TK” (my father’s initials):
I also go and fill in my mother, and my father’s brother and his daughter who connect to my 1st cousin once removed.
Next step: Those 4 red arrows on the right point to four cousins on my father’s father’s father’s side. I can fill in two more sets of ancestors:
Unfortunately, the 4 DNA matches at the right were up one generation from where they should have been.
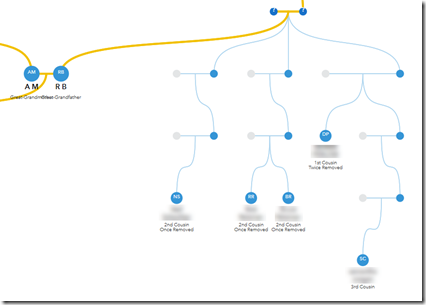
They should under AM and RB, not under RB’s parents. This is something you can’t tell from DNA, but maybe 23andMe could use ages of the DNA testers to estimate the correct generation level the matches should be at.
This is basically what 23andMe’s Family Tree beta seems to do in this, their first release. It does help visualize and place where DNA relatives might be in the tree. For example, the two unidentified cousins shown above emanate from my great-grandmother’s parents. So like clustering does, it tells me where to look in my family tree for my connection to them.
Conclusion:
This new Family Tree at 23andMe has potential. They seem to be picking specific people that would represent various parts of your tree, so it is almost an anti-clustering technique, i.e. finding the people who are most different.
There is a lot of potential here. I look forward to see other people’s comments and what enhancements 23andMe makes to it in the future, like making use of the FamilySearch relatives from their other beta. Being able to click through each DNA relative to their profile would be a useful addition. And using ages of the testers would help to get the generational level right.
Our desire as genealogists is that DNA should help us extend our family tree. It’s nice to see these new tools from 23andMe as they show that the company is interested in helping genealogists.
Now off I go to see if I can figure out how the other 7 people might be connected.












 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





