When Everything Fails At Once… - Sun, 22 Mar 2020
Remember the words inscribed in large friendly letters on the cover of the book called The Hitchhiker’s Guide to the Galaxy:
DON’T PANIC
I returned 9 days ago from a two week vacation with my wife and some good friends on a cruise to the southern Caribbean. While away, we had a great time, but every day we heard more and more news of what was happening with the coronavirus back home and worldwide.
On the ship, extra precautions were being taken. Double the amount of cleaning was being done, and purell sanitizer was offered to (and taken by) everyone when entering and leaving all public areas. The sanitizer had been a standard procedure on cruise ships for many years. I joked that this cruise would be one where I gained 20 pounds: 10 from food and 10 from purell. Our cruise completed normally and we had a terrific time. There was no indication that anyone at all had got sick on our cruise.
We flew home from Fort Lauderdale to Toronto to Winnipeg. Surprisingly to us, the airports were full of people as were our flights. None of the airport employees asked us anything related to the coronavirus and gave no indication that there was even a problem. I don’t think we saw 2 dozen people with masks on out of the thousands we saw.
After a cab ride home at midnight, our daughters filled us in on what was happening everywhere. Since we were coming from an international location, my wife and I began our at-least 2 week period of self-isolation to ensure that we are not the ones to pass the virus onto everyone else. We both feel completely fine but that does not matter. Better safe than sorry.
Failure Number 1 – My Phone
On the second day of cruise, I just happened to have my smartphone in the pocket of my bathing suit as I stepped into the ship’s pool. I realized after less than two seconds and immediately jumped out. I turned on the phone and it was water stained but worked. I shook it out as best as I could and left it to dry.
I thought I had got off lucky. I was able to use my phone for the rest of the day. All the data and photos were there. It still took pictures. The screen was water stained but that wasn’t so bad. But then that night, when I plugged it in to recharge, it wouldn’t. The battery had kicked the bucket. Once the battery completely ran out, the phone would work only when plugged in.
Don’t panic!
I had been planning to use my phone to take all my vacation pictures. Obviously that wouldn’t be possible now. I went down to the ship’s photo gallery. They had some cameras for sale but I was so lucky that they had one last one left of the inexpensive variety. I bought the display model of a Nikon Coolpix W100 for $140 plus $45 for a 64 GB SD card. I took over 1000 photos of our vacation over the remainder of our cruise, including some terrific underwater photos since the camera is waterproof.
 Before the cruise was over, my phone decided to get into a mode where it wouldn’t start up until I did a data backup to either an SD card (which the phone didn’t support) or a USB drive which I didn’t have with me.
Before the cruise was over, my phone decided to get into a mode where it wouldn’t start up until I did a data backup to either an SD card (which the phone didn’t support) or a USB drive which I didn’t have with me.
Somehow, with some fiddling, the phone then decided it needed to download an updated operating system so I wrongly let it do that. Bad move! It was obvious that action failed as then the phone would no longer get past the logo screen.
At home, Saturday at 11 pm, I ordered a new phone for $340 from Amazon. It arrived at my house on Monday afternoon and I’m back in action. The only thing on my old phone were about a month of pictures including the first 3 days of our vacation. If it’s not too expensive, I might try to see if a data recovery company can retrieve the pictures for me. If not, oh well.
Failure Number 2 – My Desktop Computer
I had left my computer running while I was gone. I was hoping for it to do a de novo assembly of my genome from my long read WGS (Whole Genome Sequencing) test. I had tried this a few months ago, running on Ubuntu under Windows. When I first tried, it had run for 4 days but when I realized it was going to take several days longer I canned it. Knowing I was going to be away for 14 days was the perfect opportunity to let it run. I started it up the day before I left and it was still running fine the next morning when I headed to the airport.
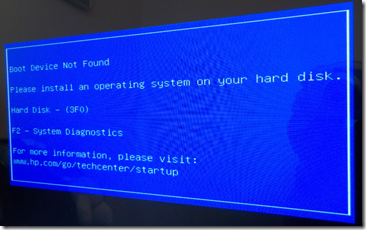
When I got back, I was faced with the blue screen of death. Obviously something happened. “Boot Device Not Found”.
Don’t panic!
I went into the BIOS and it sees my D drive with all my data, but not my C drive. My C drive is a 256 GB SSD (Solid State Drive) which includes the Windows Operating System as well as all my software. My data was all on my D drive (big sigh of relief!) but I also have an up-to-date backup on my network drive from my use of Windows File History running constantly in the background. So I wasn’t worried at all about my data. Programs can be reinstalled. Data without backups are lost forever.
I spent the rest of Saturday seeing if I can get that C drive recognized. No luck. My conclusion is that my SSD simply failed which can happen. I had a great computer but it was about 8 years old. The SSD drive was a separate purchase that I installed when I bought it to speed up startup and all operations and programs. My computer was as dead as a doorknob,
Saturday night, along with the phone I purchased at Amazon, I also purchased a new desktop at Amazon. Might as well get a slight upgrade while I’m at it. From my current HP Envy 700-209, a 4-core 4th generation i7 with 12 GB RAM, 256 GB SSD and 2 TB hard drive, I decided on a refurbished/renewed HP Z420 Xeon Workstation with 32 GB RAM, 512 GB SSD and a 2 TB hard drive for $990. It comes with 64-bit Windows 10 installed on the SSD drive. I’ve always had excellent luck with refurbished computers. The supplying company makes doubly sure that they are working well before you get them and the price savings are significant.
On Tuesday, the computer was shipped from Austin Texas to Nashville Tennessee. It went through Canada customs Thursday morning arriving here in Winnipeg at 9 a.m. and at my house just before noon.
First step, hook it up and a problem: My monitors have different cables than its video card needs. I ordered the less expensive video card with it, an NVIDEA Quadro K600. It did not come with the cables. I’m not a gamer so I don’t need a high-powered card, I made sure it could handle two monitors but I didn’t think about the cables. As it turns out, comparing my old NVIDEA GeForce GTX 645 card, I see my old card is a better card. So first step, switch my old card into my new computer.
Now start it up, update the video driver, and get all the windows updates. (The latter took about a half a dozen checks for updates and 3 hours of time)
Next turn it off and remove my 2 TB drive from my old computer to an empty slot in my new computer and connect it up. That will give me a D drive and an E drive, each with 2 TB which should last me for a while.
That was good enough for Thursday. Friday and Saturday, I spent configuring Windows the way I like it and updating all my software, including:
- Set myself up as the user with my Microsoft account.
- Change my user files to point to where they are on my old D drive.
- Set my new E drive to be my OneDrive files and my workplace for analysis of my huge (100 GB plus) genome data files.
- Reinstall the Microsoft Office suite from my Office 365 subscription.
- Set my system short dates and long dates the way I like them:
2020-03-22 and Sun Mar 22, 2020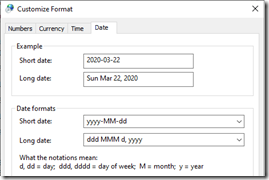
- Set up my mail with Outlook. Connect it to my previous .pst file (15 GB) containing all my important sent and received emails back to 2002.
- Reinstall and set up MailWasher Pro to pre-scan my mail for spam.
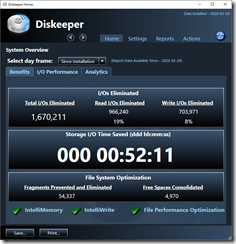
- Reinstall Diskeeper. If you don’t use this program, I highly recommend it. It defragments your drives in the background, speeds up your computer and reduces the chance of crashes. Here’s my stats for the past two days:
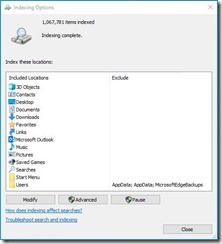
- Reindex all my files and email messages with Windows indexer:
- Change my screen and sleep settings to “never” turn off.
- Get my printer and scanner working and reinstall scanner software.
- Reinstall Snagit, the screen capture program I use.
- Reinstall UltraEdit, the text editor I use.
- Reinstall BeyondCompare, the file comparison utility I use. I also use it for FTPing any changes I make to my websites to my webhost Netfirms.
- Reinstall TopStyle 5, the program I use for editing my websites. (Sadly no longer supported, but it still works fine for me)
- Reinstall IIS (Internet Information Server) and PHP/MySQL on my computer so that I can test my website changes locally.
- Reinstall Chrome and Firefox so that I can test my sites in other browsers.
- Delete all games that came with Windows.
- File Explorer: Change settings to always show file extensions. For 20 years, Windows has had this default wrong.
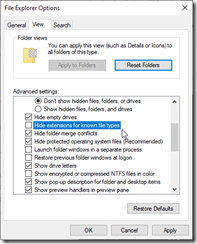
- Set up Your Phone, so I can easily transfer info to my desktop.
- Set up File History to continuously back up my files in the background, so if this ever happens again, I’ll still be able to recover.
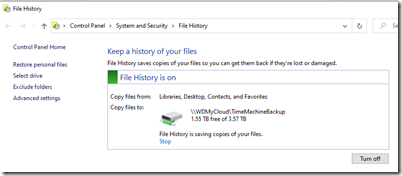
(and occasionally it saves me when I need to get a previous copy of a file) - Reinstall Family Tree Builder so I can continue working on my local copy of my MyHeritage family tree. I hope Behold will one day replace FTB as the program I use once I add editing and if MyHeritage allows me to connect to their database. I also have a host of other genealogy software programs that I’ve purchased so that I can evaluate how they work. I’ll reinstall them when I have a need for them again. These include: RootsMagic, Family Tree Maker, Legacy, PAF and many others.
- My final goal for the rest of today and tomorrow is to reinstall my Delphi development environment so that I can get back to work on Behold. This includes installation of three 3rd party packages and is not the easiest procedure in the world. Also Dr. Explain for creating my help files and Inno Setup for creating installation programs. I’ll also have to make sure my Code Signing certificate was not on my C drive. If so, I’ll have to reinstall it.
- Any other programs I had purchased, I’ll install as I find I need them, e.g. Xenu which I use as a link checker, or PDF-XChange Editor which I use for editing or creating PDF files, or Power Director for editing videos. I’ll reinstall the Windows Susbsystem for Linux and Ubuntu when I get back to analyzing my genome.
- One program I’m going to stop using and not reinstall is Windows Photo Gallery. Windows stopped supporting it a few years ago, but it was the most fantastic program for identifying and tagging faces in photos. I know the replacement, Microsoft Photos, does not have the face identification, but hopefully it will be good enough for all else that I need. Maybe I’ll have to eventually add that functionality to Behold if I can get my myriad of other things to do with it done first.
Every computer needs a good enema from time to time. You don’t like it to be forced on you, but like cleaning up your files or your entire office or your whole residence, you’ll be better off for it.
How would you cope if both your phone and computer failed at the same time?
Just don’t panic!
—Followup: After a few weeks of fiddling, I was able to get my old phone started again while plugged in, and was able to transfer my one month of photos from it to my computer via USB. So in the end, nothing important was lost.






 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





