DNA Short Snappy Opinions - Sat, 22 Aug 2020
Lots has been happening on the DNA analysis front in the past few months. Lots of very divergent opinions on a whole bunch of issues.
Here are my opinions. You are free to agree or disagree, but these are mine.
AncestryDNA
- Ancestry has had performance issues. Couldn’t they have been more honest to say performance is the reason for their cease and desist orders to the 3rd party screen scrapers who have been providing useful utilities.
- I just hate the endless scrolling screens. Bring back paging, please.
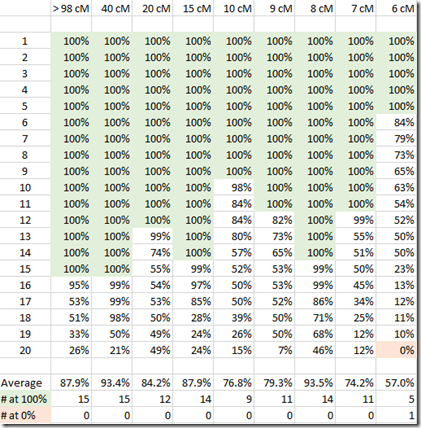
- The 6 and 7 total cM matches that Ancestry will be deleting definitely include people who have a higher probability of being related, but not because of the small DNA match which is likely false and too distant a match to ever track.
- The 6 and 7 total cM matches are also being deleted because of their performance issues.
- I in no way trust Ancestry’s Timber algorithm, especially with the longest segment length being labeled as pre-Timber to explain why it’s longer than the post-Timber total cM. Now none of their numbers make sense.
- Longest segment length is not as helpful if you have to look at it one by one. Why didn’t they show it in the match list and let us sort by it?
- Let us download our match list, please.
- Thinking Ancestry will ever give us a chromosome browser is a pipe dream.
23andMe
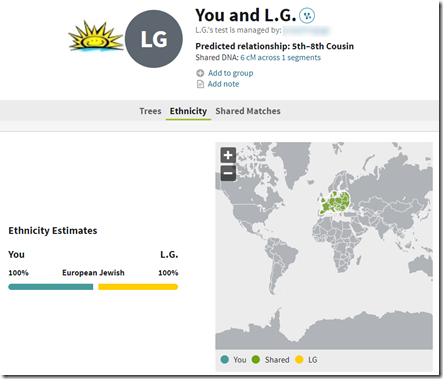
- I love that they show your ethnicity on a chromosome map. This is in my opinion, a very underutilized feature by DNA testers.
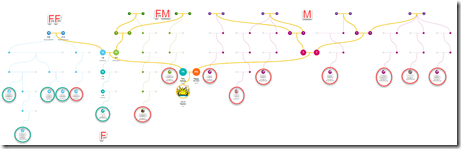
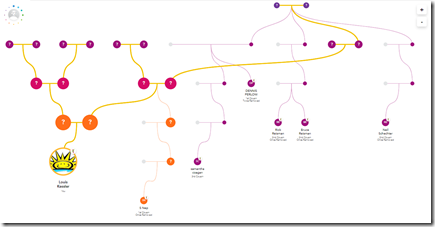
- Their Family Tree generated from just your DNA matches is a fantastic innovation.
- A month ago, some people were able to add any of their DNA matches to that family tree. They’ve never announced this and it still hasn’t rolled out to me yet. What’s the problem here? Release it, please!
- If my matches don’t opt in, I don’t want to know that. Please give me 2000 matches rather than 1361 matches that I can see and 639 that I can’t.
FamilyTreeDNA
- Lot’s of innovation that they don’t get enough credit for, e.g. their assignment of Paternal / Maternal / Both to your matches based on the Family Tree you build.
- Keeps your DNA for a looooooong time! Will be useful for future tests that don’t exist now on your relatives who passed away.
- Best Y-DNA and mtDNA analysis for those who can make use of it.
- Take advantage of their Projects if you can!
- Nobody should see segment matches down to 1 cM, or have them included in your match totals. Pick a more reasonable cutoff, please.
MyHeritage DNA
- I hate, hate, hate, did I say hate, imputation and splicing.
- As a result of the aforementioned, I believe MyHeritage has the most inaccurate matching and ethnicities of the major services.
- Showing triangulations on their chromosome browser is their best advanced feature that no one else has.
- I love that you are working with 3rd parties, and include features that others won’t such as AutoClusters.
- How about some features to connect your DNA matches to your tree, like Ancestry and 23andMe and Family Tree DNA have?
Living DNA
- They’ve missed out on a golden opportunity. They had the whole European market available.
- Three years ago they launched and promised shared matches and a chromosome browser, which they’ve still not implemented.
- Your ethnicity in no way works for me unless you add a Jewish category.
GEDmatch
- I feel so sorry for GEDmatch’s recent troubles. They are trying so hard.
- Great tools. Love the new Find Common Ancestors from DNA Matches tool that compares your GEDCOM with the GEDCOM files of your matches. Would love it more if I had any results from it.
- They let you analyze anyone’s DNA, but don’t let you download your own tool-manipulated raw data. Doesn’t that seem backwards?
- Over 100 cold cases have been solved using DNA to identify the suspect. I loved CeCe Moore’s Genetic Detective series. I can’t figure why more people won’t opt-in their DNA for police use.
ToTheLetter DNA and KeepSake DNA
- C’mon guys. We all want the stamps and envelopes our ancestors licked analyzed. This sounded so promising a couple of years ago. What’s taking so long?
Whole Genome Sequencing (WGS)
- Sorry, but today’s WGS technology will never improve relative matching the way some people think it will. Current chip-based testing already does as good a job you can do when you’re dealing with unphased data.
- Today’s WGS short read technology is too short. Today’s WGS long read technology is too inaccurate.
- The breakthrough will come once accurate long reads can sequence and phase the entire genome with a single de novo assembly (no reference required) for $100.
- PacBio is leading the way with their unbiased accurate long read SMRT technology that is not subject to repeat errors. It just needs to be about 100 times longer and remain accurate and we’re there. Optimistically: 5 years for the technology and 10 years for the price to come down.




 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





