Big update Oct 27: Much easier way to do this than in my post below. Leah Larkin informed me that I can do all 3 scenarios at once like this:
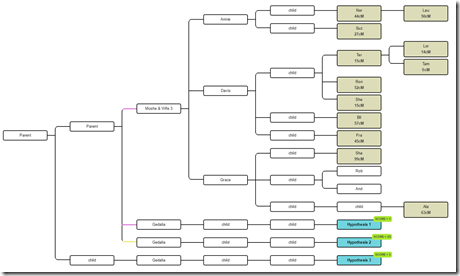
So all three hypothesis indeed can be included at once.
And the results with WATO Version 2 come out as:
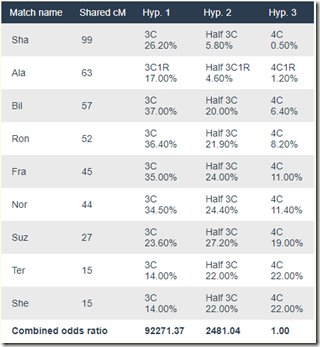
Showing Hypothesis 1 (Brother) is 37 times more likely than Hypothesis 2 (Half-Brother) which is 2481 times more likely than Hypothesis 3 (1st Cousin).
Much simpler! Many thanks to Leah and Andrew Millard on the WATO Facebook group for letting me see the light.
I’ll leave my post below to show my original thinking.
Original Post:
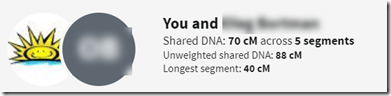
In yesterday’s post, I wanted to see if the What Are The Odds (WATO) tool at the DNA Painter site would work for endogamy, and I came out satisfied that it does, for either Ancestry DNA numbers or Family Tree DNA numbers, with the < 7 cM matches removed from the latter.
WATO is designed to help you have a DNA match with someone where you don’t know for sure how that person is related to you. You build your tree in the WATO tool and add positions where you think your match might be. You set those positions to be Hypothesis.
Well, I’ve got a slightly different problem. We’ve got a bunch of DNA matches and I know where the fit in the tree. What I don’t know is how the people at the top of the tree are related.
Let me start with the tree that I used as an example yesterday:
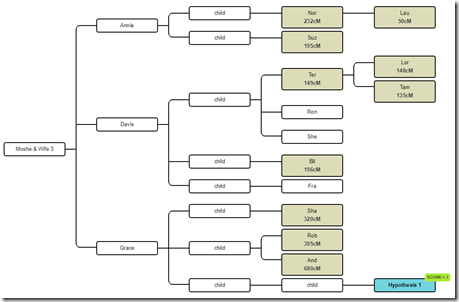
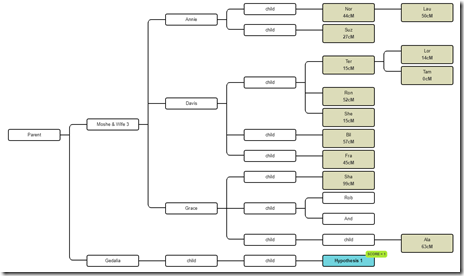
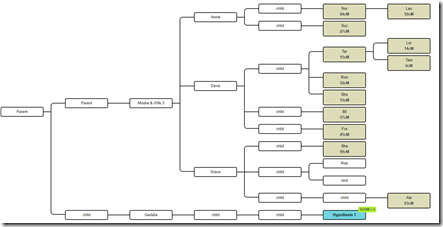
So these are all the relevant descendants of Moshe. The DNA testers are shown shaded. The Hypothesis 1 is a known tester who we simply used as a hypothesis.
Now there happens to have been a man named Gedalia who has the same last name as Moshe and came from the same town in Ukraine. We know of a few of Gedalia’s descendants who DNA tested and they are matches to the descendants of Moshe. What we don’t know and want to figure out is the relationship between Moshe and Gedalia. Could they be brothers? Half-brothers? First cousins?
Are Moshe and Gedalia Brothers?
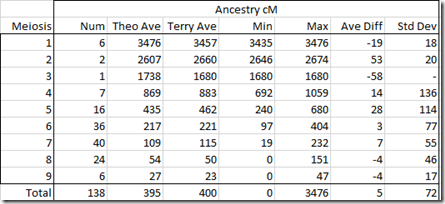
So what I’ll do is expand the tree. I’ll add Gedalia to the tree as a brother to Moshe. I’ll add the descendants and mark the one we will use in this example as the Hypothesis: Now I’ll enter the cM shared between this descendant of Gedalia and each of the testers under Moshe. I’ll used filtered Family Tree DNA numbers since those worked best yesterday:
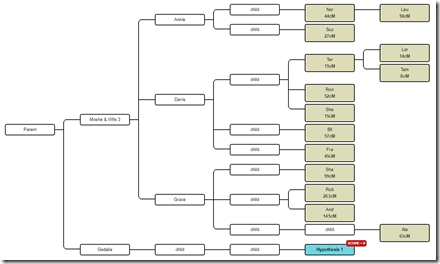
This gives us a score of zero, saying this is not possible.
So let’s take a look at the score calculation:
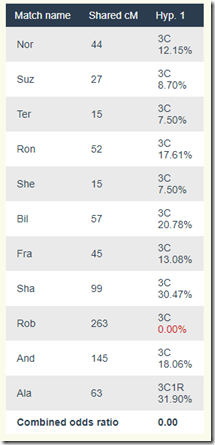
It’s saying that Rob is way too high at 263 cM to be a 3rd cousin.
But wait a minute! That is saying that Rob is related more closely than 3rd cousin to our Hypothesis person, who we’ll call: Hyp. We know from the diagram above that through Moshe and Gedalia, he cannot be closer than 3rd cousins. Since Rob’s cousin Sha and 1C1R Ala don’t have the same problem, they are okay. That must mean that Rob’s mother is related to Hyp, adding extra cMs to Rob and his sibling And. In fact, And is higher than all the rest at 145 cM, but not high enough to make being a 3rd cousin to Hyp an impossibility.
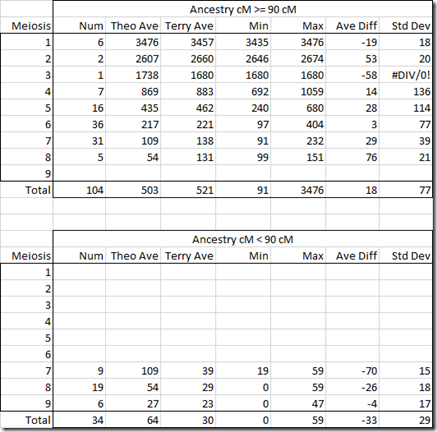
Since Rob and And are related another way to Hyp, what I’ll do is remove their shared DNA amounts from being included in the WATO calculations and run it again:
That’s better and now the Hypothesis shows up as possible. Here’s the score calculation:
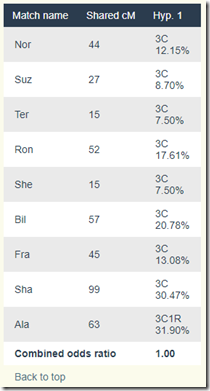
It’s the same as the above for the listed people, except that the Combined odds ratio is now 1.00.
Are Moshe and Gedalia Half-Brothers?
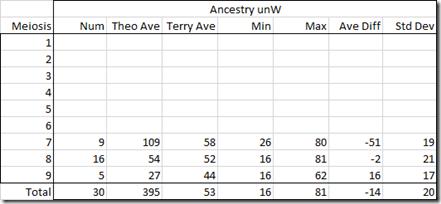
Let’s now do the same thing and just change Moshe and Gedalia to be half-brothers. WATO lets us do this and indicates they are halves with the coloured dotted lines to the left of their boxes:
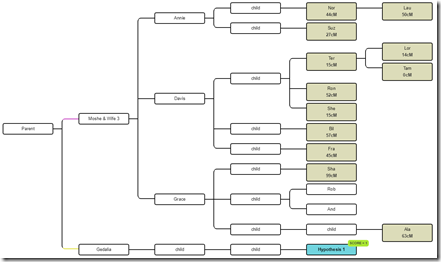
All of the scores have changed, but this scenario is still a possibility:
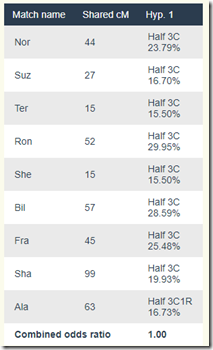
Are Moshe and Gedalia First Cousins?
Well, let’s delete Gedalia’s side and add him back in as a first cousin:
Once again, this is said to be possible. Here are the scores:
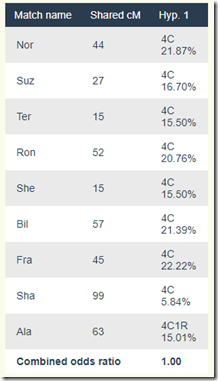
So Which Is More Likely? Brother? Half? Cousin?
WATO has a wonderful mechanism for comparing different Hypotheses. When you include more than one hypothesis in a scenario, it tells you which of the three is most likely and how many times more likely it is than the next. (See yesterday’s post for an example).
But here, I have three different trees each with only one Hypothesis. WATO won’t compare them for you.
Well I think I see what WATO is doing. I may be wrong, but it looks like it is multiplying the probabilities together and comparing the results between the scenarios. So I can easily do that myself in a spreadsheet:
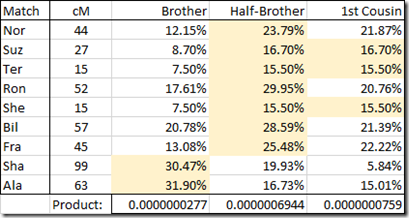
I have highlighted the most likely scenario for each match. Half-Brother wins this comparison with 7, versus 1st Cousin with 3 and Brother with just 2.
The line at the bottom contains the product of the 9 values above it. The highest value is Half-Brother which is 9 times larger, meaning it is 9 times more likely a possibility than 1st Cousin. 1st Cousin is 3 times more likely than Brother. And Brother is 25 times less likely than Half-Brother.
So there you have it. We haven’t proved anything, but at least we now know that all scenarios are possible and that half-brother is most likely.
Hint, Hint, Leah and Jonny
WATO is a wonderful tool to help you hypothesize where your DNA matches fit into your tree. That was what it was designed for.
But wouldn’t it be nice if WATO could also help you test different ancestral scenarios as well, as I have just done? Well it can, if you follow the above procedure and do the comparison yourself,
WATO-Ancestors could be set up to make it easier for you by remembering the results of each of your scenarios, and then comparing them for you, so that you won’t have to yourself.
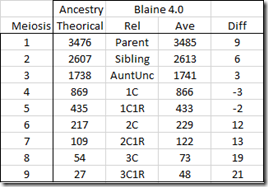
Update (80 minutes later): I didn’t realize when I was doing the analysis that I was using Version 1 of WATO. Version 2 includes new probability numbers taken from an update to Ancestry’s paper. See Leah’s article: Improving the Odds. The main improvement is that it now has much more detail for small matches.
You can switch from Version 1 to 2 very easily, so I did and I recalculated. Here’s the revised table:
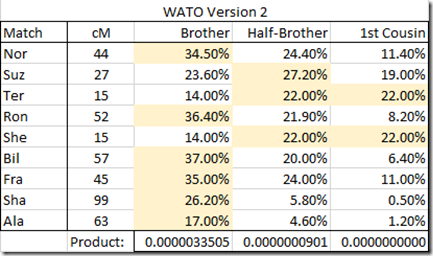
To tell the truth, it really changed the results. Now the conclusion is that Brother is the most likely relationship and that scenario is 37 times more likely than Half-Brother.
So make sure you use Version 2 of WATO to get the best probabilities.
Additional Idea: If you have more than one tester on the other side of the tree, you can calculate all the match values for each scenario for each of them, and then simply multiply out (or geometric mean) the “Product” line for each of them.
For example, in the above table, if I had a second person that gave Product numbers of 0.0000385 for Brother, 0.0000655 for Half-Brother and 0.0000073 for 1st Cousin, then
GMean(Brother) = (0.0000033505 * 0.0000385) ^ (1/2) = 0.0000114
GMean(Half-Brother) = (0.0000000901 * 0.000655) ^ (1/2) = 0.0000024
GMean(1st Cousin) = (0.0000000 * 0.0000073) ^ (1/2) = 0.0000000
If you don’t know what a geometric mean is, then just use a simple average which should still tell you which scenario is most likely.















 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





