Catching Up - Sat, 8 Oct 2022
I was shocked to notice that I hadn’t blogged in over 3 months. I think it’s time to catch up with what’s been going on.
The Last 3 Months
To be honest, I’ve had another non-genealogy project distracting me and taking some of my genealogy/programming time away. Also, we had a beautiful summer here in Winnipeg, and I’ve been doing lots of bike riding and swimming. It’s much harder to be indoors on my computer for programming when its so nice outside.
Double Match Triangulator
I’ve actually released 6 minor versions (5.0.1 to 5.0.6) since June. Most have had to deal with changes at GEDmatch. They seem to have been making a lot of improvements there and their changes can break DMT. I always try to address anything no longer working as soon as I find out about them.
Meanwhile, I have been involved for over a year on a DNA project with my wife’s 3rd cousin. He had close to 60 descendants of my wife’s great-grandfather and five suspected siblings, and we are trying in spite of endogamy to determine whether they are indeed siblings. I have been trying to to use DMT to help with the analysis and the data has been an excellent test bed for DMT. It has led to improvements included in Version 5.0 that was released in May.
I still have some work to do on that study, and that may test DMT some more.
Behold
During the Spring, I was able to spend a couple of months working on Behold, trying to finish up Version 1.3 and get the Everything Report to be how I wanted it. But then the DMT work and the summer got in my way.
For the longest time, I’ve always expected Behold would become the genealogy editor that I would use for recording my genealogy. But I never got past it being just a genealogy data viewer. A few years back, In 2018, I decided I couldn’t wait for myself and Behold any more and I needed an editor for my genealogy. I decided to use MyHeritage online and its Family Tree Builder software on my computer. I am very happy that I did.That’s because there’s nothing like the smart matches that do a lot of research for you, pointing at records that more often than not are correct, and connecting you with other researchers and their trees.
As a result, my ideas for Behold have changed. I now have all my family tree data up on MyHeritage, and some of my data up on Ancestry, FamilySearch, WIkiTree, Geni, Geneanet and GenealogyOnline. Of those, FamilySearch, WikiTree and Geni are one-world trees, meaning other people are editing and updating my family there as well.
What I need now, rather than an editor, is a tool to help me evaluate and compare the information I have on MyHeritage to the information that’s on the other trees. I need to be able to find out what’s changed from my own information so that I can determine what needs to be updated. And I’d like some help to keep the thousands of profiles I have on the various systems in sync along with the photos and records attached to them.
There used to be a program called AncestorSync. It was designed to keep all your trees in sync on the various systems. It would have been perfect. But unfortunately, the team stopped developing the program.
I have no desire to try to get Behold to actually Sync your data. That would be a lot of work as I’d have to make agreements with the various tree operators to be allowed to update to their systems, and I’d have to be meticulous to learn to use their APIs (Application Program Interface) to make the update without mistakes. That’s a bit more than I want to take on.
Instead, I could do the next best thing: Set up Behold to help you manually update another tree with your data from a different tree. I know I myself want that function, and I would think that a lot of other people might find that very useful as well. We’ll see how it goes.
Conferencing and Social Media
After over 2 1/2 years of our Covid world, in-person genealogy conferences are finally starting up again. During the 2010’s, I attended and gave talks at 11 International Genealogical Conferences, 6 in the United States, 3 in Canada, 2 on the high seas around Australia and New Zealand, and 1 in The Netherlands.
2020 changed things and the world started going virtual. Now we’re inundated with a cornucopia of genealogical webinars available to us while sitting at home in our pajamas on our computer. I know I have watched hundreds of genealogy presentations in the past 30 months through providers such as Legacy Family Tree Webinars, the Virtual Genealogical Association, RootsTech, Dear Myrtle, Family History Fanatics, WikiTree, GeneaBlogger, Ed Thompson, the Association of Professional Genealogists, and presentations by the FGS and local genealogy societies – not just where I live but from all over the world. I have also been a speaker at several of these online conferences.
And last year I took an excellent online SLIG (Salt Lake Institute of Genealogy) course to help me with my Russian research.
I have got to the point where my head is 98% full and I’ll be starting to be more selective of what webinars I watch and conferences I go to. There will have to be something quite new and appealing to attract me now.
Social media is sort of the same thing. Genealogy groups on Facebook exploded in popularity in the past 5 years. I participated quite a bit at the beginning, but they have become less useful to me since then.
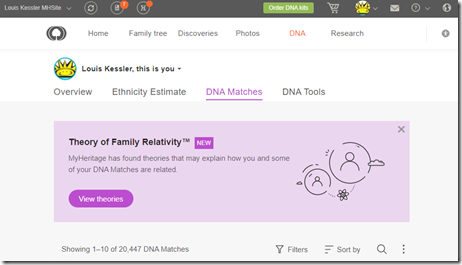
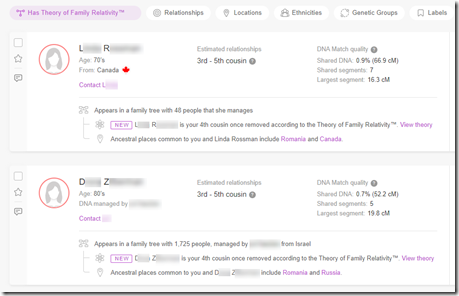
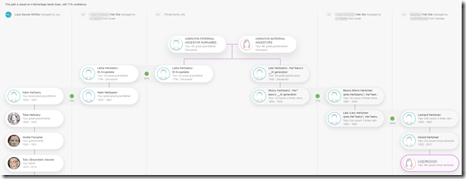
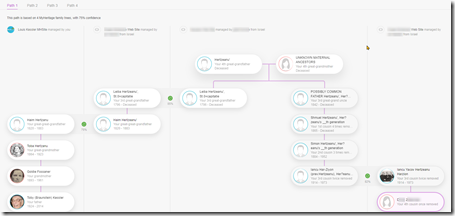
DNA
I’ve done just about everything I can with my own DNA. I’ve tested at all the sites including Big-Y and mtDNA at Family Tree DNA. I’ve used all the tools at all the sites as well as many 3rd party DNA tools. Endogamy and ancestry that only goes back only to the early 1800’s (Romania and Russian Empire) has limited my ability to connect to many of my DNA relatives. I have even taken a few WGS (Whole Genome Sequencing) tests to see what they could do for me.
Looking to the Future
My genealogy has taken great steps in the past couple of years, due to the help of MyHeritage hints and also hundreds Russian and Romanian records of my family found for me by some excellent researchers. I’ve got to get this all organized and fully documented.
I’ll be continuing to work on Behold to help me with this, and will update DMT as needed. I’ve got my GenSoftReviews site that I’ll keep maintaining as well as my personal lkessler.com website. Maintaining websites, by the way, does take a fair bit of time. Especially when an issue like updating a PHP version takes hold.
I’m still very excited about the future, because we never know what’s in store.

 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





