Evaluating Ancestry DNA’s Parental Matches - Mon, 26 Dec 2022
A couple of months ago, Ancestry came out with a new feature called Parental Matches, that shows which parent a DNA match is connected to.
My own DNA should be a good test for this. I have 100% endogamy on all my ancestral lines, but my parents are not related according to GEDmatch’s test.
Both my parents passed away before I started DNA testing, but I did test my uncle (my father’s brother) at FTDNA and I have some first cousins who tested at Ancestry and other sites, so I can use them for additional comparisons.
Ancestry’s Parental Matches stems from the SideView technology introduced about 6 months earlier that reveals your ethnicity inheritance from each parent. The SideView ethnicity breakdown really didn’t help me very much:
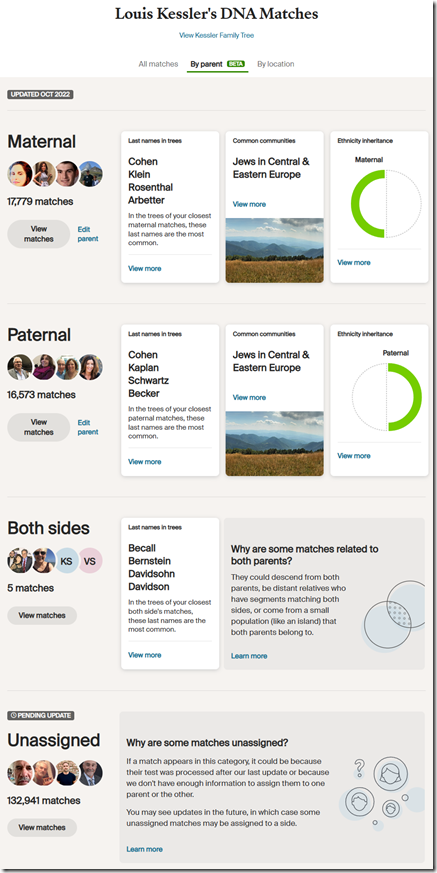
Ancestry DNA’s Parental Matches
Of my 167,298 matches at Ancestry DNA, I have:
- 17,779 matches (10.6%) that are assigned Maternal.
- 16,573 matches (9.9%) that are assigned Paternal.
- Only 5 matches assigned to Both sides.
- 132,941 matches (79.5%) that are Unassigned.
I am somewhat surprised that more of my mother’s side is assigned than my father’s, since I have more known relatives on my father’s side in both my family tree and among my DNA matches.
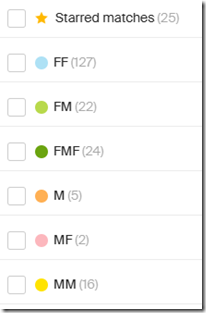
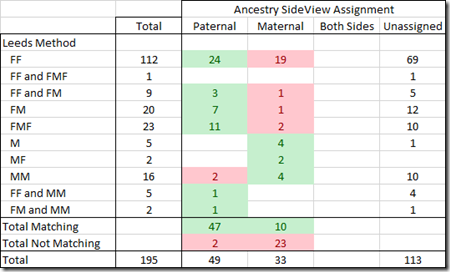
I have in the past used the Leeds Method on my Ancestry matches with reasonably good success. That allowed me to assign is most cases one grandparent and in a few cases more than one grandparent to my closest matches at Ancestry. I assigned each match one or more colored dots representing the grandparent(s):
Where FF = my Father’s Father’s side, FM = my Father’s Mother’s side, etc.
Most of the Leeds assignments are on my Father’s side.
- 173 on Father’s sides, made up of 127 on Father’s Father, 22 on Father’s Mother, and 24 on Father’s Mother’s Father
- 23 on Mother’s sides, made up of 2 on Mother’s Father, 16 on Mother’s Mother and 5 on Mother’s side but grandparent unknown.
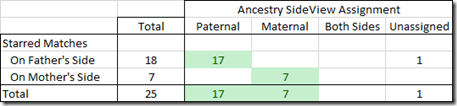
The 25 starred matches are the people that I know how I am connected to and that I have in my family tree.
So the question is, how well does Ancestry’s Parental assignments match both my known relationships and my Leeds assignments.
For my known relationships, it did pretty good:
It assigned 24 of my known relatives correctly and the other was not wrong, but was just not determined.
My assumption would be that Ancestry must be using my Ancestry family tree itself to make the assignments. Otherwise it would not be able to determine what side any of my relatives are on. The one unassigned match might have been left that way because being a 4th cousin, he is my most distant known DNA match, and maybe Ancestry does not use the tree for cousins that distant.
How well do the assignments match the Leeds method?
Of my DNA matches where Ancestry assigned a parent, it agreed with the Leeds Method in 57 out of 82 cases (70%). So Ancestry and Leeds disagree about 30% of the time. Unfortunately, we cannot in this case tell which one is incorrect.
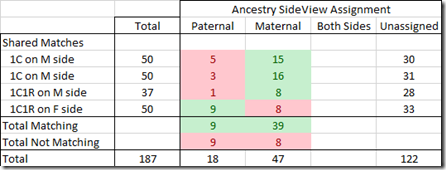
Comparing Shared Matches
At Ancestry, I have DNA matches with two 1st cousins on my mother’s side, with one 1C1R on my mother’s side and one 1C1R on my father’s side. My shared matches with each of them should be expected to mostly match only people on the same side as they are.
If I look at the first 50 shared matches with each of them, this is what I see. (My 1C1R on my mother’s side only has 37 shared matches)
For shared matches, 48 out of 65 cases (74%) match the side the cousin is on. That’s not bad, but is a bit less accuracy than I would have hoped for since my parents are not related.
Side Assignment at Family Tree DNA
Family Tree DNA has had Paternal and Maternal matches assigned to each side for a long time. Like Ancestry, since I don’t have parents tested, it assigns sides based on comparison with testers that who I’ve placed in my family tree there.
I really only have two close relatives in my tree at FTDNA, my uncle (father’s brother) and the same “1C1R on M side” as at Ancestry.
With the two of them in my tree and a couple more distant relatives, FTDNA assigns my 35,169 matches as follows:
- 1,564 matches (4.4%) that are assigned Maternal.
- 10,732 matches (30.5%) that are assigned Paternal.
- 421 matches (1.2%) that are assigned to Both sides.
- 22,452 matches (63.9%) that are Unassigned.
Compare this to Ancestry’s assignments. There are so many more Paternal assignments at FTDNA than maternal, but Ancestry’s assignments are about 50:50. I don’t know why this is.
If I look at my first 50 matches in-common-with my uncle, 29 are assigned Paternal and only 2 are assigned maternal. That’s 94% correct.
With my 1C1R, 6 are assigned Maternal, but 15 are assigned Paternal. That’s only 29% correct. Again, I don’t know why.
It might be a bit of a Paternal bias at FTDNA. I don’t know if this is real (because of more people on my Paternal side testing there) or if it is a by-product of the way their algorithms work with my matches.
Also, I don’t know why I have 6009 matches in common with my 1C1R at FTDNA but only 37 in common at Ancestry.
Conclusion
Parent assignments on your DNA matches is a nice feature to have, especially when you haven’t had your parents tested.
Family Tree DNA has had this for a while using information from the family tree you put together on their site. Ancestry has now just added this.
The assignments appear to be fairly good and seem to be correct more often than not. They are useful, but should be thought of as a reasonable hint, rather than an accurate determination.
There are more advanced techniques that the companies could use to make this feature more accurate and more useful. Incorporating automated Leeds assignments, clustering techniques, or comparing segment matches would improve the results.
23andMe assigns parent information only if you have parents tested. MyHeritage DNA, Living DNA and GEDmatch do not yet assign parent sides to your matches.
Parental assignment is a good feature. The companies should all include it, and be working to make it more accurate and useful.
Followup: Jan 4, 2023: Family History Fanatics (Andy Lee) just posted a YouTube video worth watching: How ACCURATE is Ancestry’s Parent1/Parent2 DNA Match Separator? Andy found 90% of his DNA matches had a parent assigned compared to only about 20% for me. He also had very few matches with “Both” parents assigned.
That got me to go back and look at my matches again, and I was surprised to see that only a week later, my numbers changed to:
- 18,823 Maternal (up 1044)
- 17,038 Paternal (up 465)
- 963 Both sides (up 958)
- 130,795 Unassigned (down 2146)
- 167,619 Total (up 321)
So a couple thousand more assignments were made in the last week, and my very low number of Both Side (5) was substantially increased to 963.







 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





