To my surprise, I downloaded my raw data from my 23andMe DNA test and it was different from my earlier downloads.
I would have thought your raw data from a company wouldn’t change. I took one DNA test there, so my results should be determined once, and that’s what should be represented in my raw data. I don’t care if the format of the file changed, but I do care if the data represented in that file changed.
So lets see what might have happened here.
Three 23andMe Raw Data Downloads
I took my 23andMe test in Nov 2017. I’ve downloaded my raw data 3 times since then.
I talked about my original download in my August 2018 article Comparing Raw Data from 5 DNA Testing Companies. My April 2020 article Determining the Accuracy of DNA Tests used my 2nd download. I noticed a difference in the counts from my 23andMe file from and the earlier article, and I said at the time “I’m not sure why there’s a difference” and I assumed I must have made some mistake with the numbers. But what I see now is that the files were slightly different.
Let’s compare the counts in the 3 files I now have:
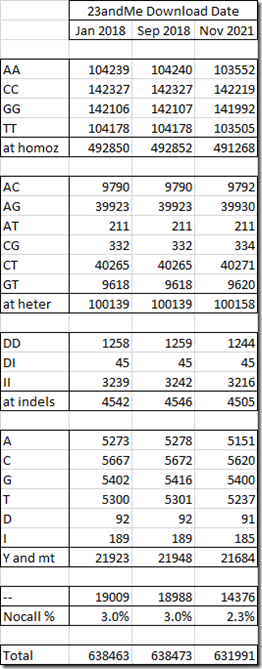
Above are the counts of the SNP values in groups: the autosomal homozygous (same valued) SNPs, the autosomal heterozygous (different valued) SNPs, the autosomal Insertions and Deletions, the Y and mt chromosomes which have just one value, and the no-calls (values which could not be determined with sufficient accuracy that are shown as a double dash “- -“).
Comparing the counts between the first two downloads, we see they are fairly close with the total number increasing by 10 and the largest difference only being 14 in the G group.
But you can see my most recent download had a significant change, with the total number of SNPs decreasing by almost 6500. About 4500 of those was from a reduction in the number of no-calls, but the other 2000 were because of fewer actual values. Surprisingly, the number of autosomal heterozygous SNPs went up by 19.
So What’s Changed?
Since I created my “All 6” Combined Raw Data File in my May 2019 article Creating a Raw Data File from a WGS BAM file, and that was based on my Sep 2018 download from 23andMe, I’ll compare that download to the one I just did.
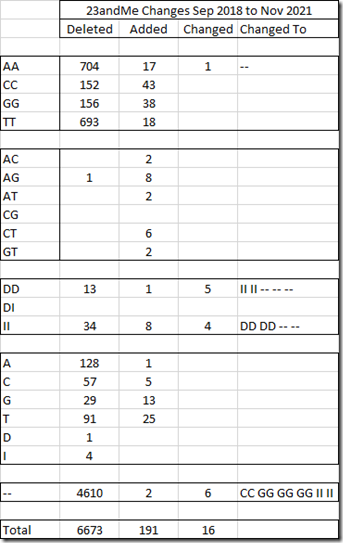
So there were 6673 SNPs in my old file that are not in my new file. 4610 of those were no calls in my old file, so those don’t matter. And 52 were deletions or insertions that other companies don’t even report. But that still leaves 2010 SNPs that had values previously and no longer do. Of those, 1706 are autosomal SNPs that are important for DNA matching.
There were 191 new SNPs in my new file that were not in my old file. And 136 of those are autosomal SNPs that are important for DNA matching.
And there were 16 SNPs that changed values in my new file. Fortunately none of the changes were important. There were 6 nocalls changed to values and 9 values that were either changed to nocalls or insertions or deletions.
Did We Lose Useful Information?
The question is whether we lost useful information in those 2010 deleted SNPS that had values previously or if we gained useful information from those 182 new SNPs.
To find out, I have to go to my April 2020 article Determining the Accuracy of DNA Tests. See the section about the Accuracy of Standard Microarray DNA Tests. If I do the same procedure and compare the values from the 4 BAM files that agree with each other to these deleted and added SNPs, I can get an approximate accuracy estimate for them.
Of the 1706 autosomal non-indel SNPs that were in the old 23andMe file, 1518 had identical values in the 4 BAM files. Of those, only 1164 match the deleted value. That’s an error rate of 23.3% which isn’t good at all. So what we lost were SNPs with a high error rate.
Of the 136 new autosomal SNPs, 129 had identical values in the 4 BAM files. Of those, 128 matched the new 23andMe value. Just one, which was AG in the 23andMe file and was AA in the BAM files didn’t match. That’s an error rate of 1 / 129 = 0.8% which is okay. So what was added was useful.
The values deleted had a high error rate. The values added had a low error rate. I don’t know the reason why 23andMe made these changes or what they did to make the changes, but the net result was a slight overall improvement to the accuracy of their raw data file.
Did the Raw Data File of Other Companies Change as Well?
Let’s see.
Family Tree DNA: My first build 37 raw data file download was from Aug 2018. My download today is identical to it.
Ancestry DNA: My first raw data download was Mar 2018. Compared to my download today, 56 SNPs from the early file have been deleted. All the deleted SNPs had values and none were no-calls. No SNPs were added and no SNPs were changed. So this is a change, but a minor change.
LivingDNA: My first raw data download was Aug 2018. My download today is identical to it.
MyHeritage DNA: My first raw data download was March 2017. But I was surprised to find my download today is very different, much more different than the 23andMe data, and needs its own analysis, which follows.
MyHeritage DNA Raw Data Changes
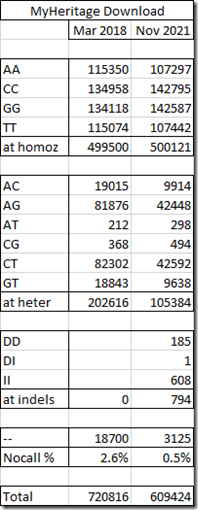
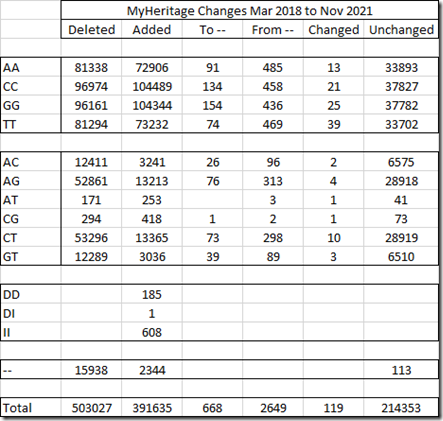
Here is my MyHeritage DNA comparison table:
There are over 110,000 fewer SNPs in the new dataset from MyHeritage. Most of the reductions appears to be among the heterozygous SNPs which halved in numbers.
I hadn’t heard that any change was made to MyHeritage DNA’s raw data files, so I didn’t expect this. I also see they added a few indels to their file, similar to the way 23andMe does and have cut down the number of no-calls.
But just take a look at these changes:
Of the 720,816 SNPs they had in the original file, they only retained 214,353 of them, changed 3436 of them, and added 391,635 new SNPs that weren’t there before.
This is a major change! The original MyHeritage raw data I got in 2018 is nothing like the new one I now get. The SNPs they are using now are 65% different.
Hopefully they improved their accuracy. Let’s see.
When I wrote my Determining the Accuracy of DNA Tests article in Apr 2020, I produced the following table:
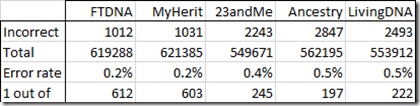
My numbers showed that MyHeritage had the best accuracy of the 5 companies, just 1 error out of every 603 SNPs.
When I do the same comparison of my new MyHeritage DNA raw data, I get this:
My new data has 574,057 autosomal values on Chr 1 to 22 that are not no-calls. Of those, 534,179 have agreeing values in my 4 BAM results that I can compare them to. 529,707 of those match the MyHeritage value.
That means 4472 are incorrect out of 534,179 or 0.8%. So 1 out of 119 values in my new MyHeritage raw data file are incorrect. It’s error rate has increased by a factor of 5.
With this change, MyHeritage went from being the most accurate of the 5 companies, to being the least accurate.
I don’t know exactly when or why MyHeritage made changes to what it puts in your raw data download file, but whatever they did (maybe their imputation and splicing) decreased the quality of its raw data considerably.
I cannot say for sure what that does to MyHeritage’s matching accuracy. That will depend on their matching algorithm and whether they are allowing for the possibility that 1 out of 100 SNPs may have an incorrect value, rather than the 1 in 600 that they had before. If they did compensate for this and lessened their requirement to, say, 1 mismatch every 50 SNPs, then you will have more false segments than you did before.

Joined: Sat, 17 Aug 2019
3 blog comments, 0 forum posts
Posted: Sat, 20 Nov 2021
Very nice write up. Thanks!
Do you think it’s possible that some of the changes you observe are because of changes in the underlying rsid SNP database? I have in my own research sometimes stumbled on SNP rsids that where merged or renamed. Do you make your comparisons based on rsid or position?
Joined: Sat, 17 Aug 2019
3 blog comments, 0 forum posts
Posted: Sat, 20 Nov 2021
Oh and what about genome versions, do they differ?
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 21 Nov 2021
Yinwang: I compared both by RSID and by position. In one comparison between two files, for example, I noted to myself that 299 RSIDs had changed at the same position, and that 3 RSIDs had different positions. But mentioning that in the article would have been off-topic and would only have complicated what I was trying to say. My final comparison was by position, because we were worried about the value there, and didn’t care about how they were identifying the SNP via the RSID.
Regarding genome versions, all the company downloads are using Build 37, so theoretically everything should be compatible between them, or at least within one company. But the difference in RSID to positions I mention above do indicate something is off-kilter and the companies are making small changes to their reference genome from time to time.
Joined: Tue, 30 Nov 2021
1 blog comment, 0 forum posts
Posted: Tue, 30 Nov 2021
Have you looked closely at the y DNA data from 23andme or ancestry to see how often those snps have changed in a material way? Those would have greater consequences for genealogy where folks are leaning in those results to compare haplogroups. Thanks for the interesting post!