In my last blog post, I analyzed my segment matches at 23andMe with Double Match Triangulator,. This time let’s do the same but with my GEDmatch segment matches.
Getting Segment Match Data from GEDmatch
At GEDmatch you need their Tier 1 services (currently $10 a month) in order to download your segment matches. But you can download anyone’s segment matches, not just your own. But+But they don’t include close matches of 2100 cM or more, meaning they won’t include anyone’s parents, children, siblings, and maybe even some of their aunts, uncles, nephews or nieces. The But+But could in some cases be problematic because people who should triangulate will not if you don’t include their close matches. But+But+But even that should still be okay in DMT since DMT’s premise is to use the matches and triangulations that exist with the ideas that there will generally be enough of those to be able to determine something.
GEDmatch not too long ago merged their original GEDmatch system and their Genesis system into one. Now all the testers on GEDmatch who used to be in two separate pools, can all be compared with each other. While doing so, GEDmatch also changed their Segment Search report now providing a download link. The download is in a different format than their screen report is and used to be. With all these changes, if you have old GEDmatch or Genesis match file reports that DMT helped you download, you should recreate each of them again in the new format. Check DMT’s new download instructions for GEDmatch.
When running GEDmatch’s Segment Search, the default is to give you your closest 1000 kits. I would suggest increasing that to give you your 10000 closest kits. I’ve determined that it is definitely worth the extra time needed to download the 10000 kits. For example, in my tests, comparing 1000 kits versus 1000 will average about 50 people in common. Whereas comparing 1000 kits versus 10,000 kits will average 350 people in common. So 300 of the 1000 people in common with Person A are in not in the first 1000 kits in common with Person B, but are in the next 9,000 kits. When using the 1000s, DMT can cluster 18% of the people. When using 10,000s, that number goes up to 56%
It can take anywhere from a few minutes to an hour to run the 10,000 kit Segment Search at GEDmatch, so if you have 10 kits you want to get segment matches for, it could take the good part of a day to complete.
I downloaded the segment match data for myself and 8 people I match to using the 10,000 kit option. They include 3 relatives I know, and 5 other people who I am interested in.
My closest match is my uncle who shares 1958 cM with me. GEDmatch says:
I really don’t know why GEDmatch does this. To find the one-to-one matches of a few close relatives and include them in the segment match list would use only a tiny fraction of the resources required overall by their segment match report, The penalty of leaving out those close matches is huge as matches with all siblings and some uncles/aunts, nephews and nieces are left out. Parents and children are also left out, but they should match everywhere.
I have added into DMT an ability to download the one-to-one matches from GEDmatch for your matches that the Segment Search does not include. In my case, my uncle is 1958 cM, so I didn’t need to do this for him. You can also use the one-to-one matches to include more distant relatives who didn’t make your top 1000 or 10000 people.
Entering my Known Relatives’ MRCAs in my People File
These are the 3 people at GEDmatch that I know my relationship to and who is our Most Recent Common Ancestor (MRCA).
- My uncle on my father’s side, MRCA = FR, 1958 cM
- A daughter of my first cousin on my mother’s side, MRCA = MR, 459 cM
- A third cousin on my father’s mother’s father’s side, MRCA = FMFR, 54 cM
(The F, M, and R in the MRCA refer to Father, Mother and paiR of paRents. So an MRCA of MR are your mother’s parents. FMFR are your father’s mother’s father’s parents. MRCAs are always from the tester’s point of view.)
Since DNA shared with my uncle can come from either of my paternal grandparents, and since DNA from my 1C1R can come from either of my maternal grandparents, their double matches and triangulations do not help in the determination of the grandparent. However, they should do a good job separating my paternal relatives from my maternal relatives.
The third cousin will allow me to map people to my FM (father’s mother) grandparent and to my FMF (father’s mother’s father) great-grandparent.
Lets see how this goes.
Painting
At GEDmatch, I only have the one third cousin to work with to determine grandparents. My cousin only shares 54 cM or about 1.5% of my DNA. The process DMT uses of automating the triangulations and extending them to grandparents, then clustering the matches and repeating, results in DMT being able to map 44% of my paternal side to grandparents or deeper. This is using just this one match along with my uncle and my 1C1R.
Loading the mappings into DNA Painter gives:
If you compare the above diagram to the analysis from my 23andMe data I did in my previous post, you’ll see a few disagreements where this diagram is showing FM regions and the 23andMe results show FF regions. These estimates are not perfect. They are the best possible prediction based on the data given. If I was a betting man, I would tend to trust the 23andMe results more than the above GEDmatch results in these conflicting regions because 23andMe had many more MRCAs to work with, including some on both the FF and FM sides, than just the one FMF match I have here at GEDmatch.
The bottom line is the more MRCAs you know, the better a job DMT can do in determining triangulation groups and the ancestral segments they belong to. None-the-less, using only one MRCA that specifies a grandparent, this isn’t bad.
Clustering
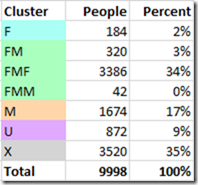
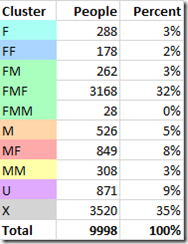
DMT clusters the 9998 people in my GEDmatch segment match file as follows:
DMT clustered 39% of the people I match to as paternal and 17% as maternal.
34% were clustered into one big group, my Father’s Mother’s Father (FMF) which is higher than the expected percentage (12.5%). My third cousin whose MRCA is FMFR might be biasing this a little bit. Every additional MRCA you know will add information that DMT can work with to help improve its segment mappings and clustering. I just don’t happen to know any more at GEDmatch, so these are the best estimates I can do from just the GEDmatch data. As more people test, and I figure out how some more of my existing matches are connected, I should be able to add new MRCAs to my GEDmatch runs.
Grandparents on My Mother’s Side
I have one relative on my mothers side included here at GEDmatch. Being the daughter of my first cousin, she is connected through both my maternal grandparents. Her MRCA is MR and DMT can’t use her on her own for anything more than classifying people as maternal.
I don’t like the idea of being unable to map grandparents on my mothers side. I don’t have any MRCAs to do that.
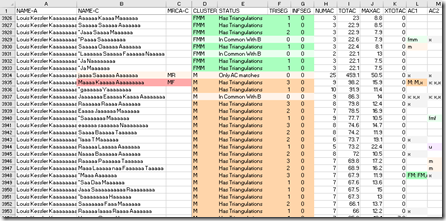
But I have an idea and there is something I can try. If I take a look at the DMT People File and scroll down to where the M cluster starts, you’ll see my 1C1R named jaaaa Saaaaaa Aaaaaaa with her MRCA of MR. Listed after her are all the other people that DMT assigned the cluster M to and they are shown by highest total cM.
The next highest M person matches me with 98.2 cM. That is a small enough number that the person will be at least at the 2nd cousin level, but more likely the 3rd or 4th cousin level.. Since they are further than a 1st cousin, they should not be sharing both my maternal grandparents with me, but should only share one of them. I don’t know which grandparent that would be, but I’m going to pick one and assign them an MRCA to it. This will allow DMT to distinguish between the two grandparents. I’ll just have to remember that the grandparent I picked might be the wrong one.
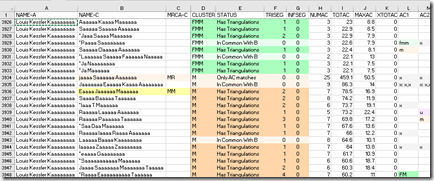
So I assign the person Maaaa Kaaaaa Aaaaaaaaa who shares 98.2 cM with me the MRCA of MF as shown below. Note I do not add the R at the end and make it MFR. The R indicates the paiR of paRents, and indicates you know the exact MRCA and share both of those ancestors. DMT accepts partial MRCAs like this.
DMT starts by assigning the MRCA you give it. Unlike known MRCAs whose cluster is always based on the MRCA, DMT could determine a different cluster for partial MRCAs.
I run DMT adding the one partial MRCA to this one person. Without even downloading the segment match file for Maaaa Kaaaaa Aaaaaaaaa, DMT assigned 976 of the 1674 people that were in the M cluster to the MF cluster, leaving the other 698 people in the M cluster. In so doing, it painted 11.5% of the maternal chromosome by calculating triangulations with Maaaa Kaaaaa Aaaaaaaaa and then extending the grandparents based on other AC matches on the maternal side that overlap with the triangulations.
That’s good. Now what do you think I’ll try. Let’s try doing it again. Likely many of those 698 people still assigned cluster M are on the MM side. So let’s take the person in the M cluster with the highest cM and assign them an MRCA of MM. But we have to be a bit careful. That person with 86.3 cM named Jaaaaaaa Eaaaaa Kaaaa Aaaaaaaa has a status of “In Common With B”. That means they don’t triangulate with any of the B people. If they don’t triangulate, DMT cannot assign the ancestral path to others who also triangulate in the same triangulation group at that spot. So I’ll move down to the next person named Eaaaa Jaaaaaa Maaaaaa whose status is “Has Triangulations” and shares 78.5 cM and assign the MRCA of MM to her, like this:
After I did that, I ended up with 296 people assigned the MM cluster, 859 assigned MF (so 117 were changed), and 519 still left at M. meaning there were still some people that didn’t have matches in any of the triangulation groups that DMT had formed, or maybe they had the same number of MF matches as they do MM matches so no consensus. Or maybe the people I labelled MM and MF are really MMF and MFM and these people left over are MMM or MFF thus not triangulating with the first two. It could be any of those reasons, or maybe their segment is false, I don’t know which. In any case, I now have 23% of my maternal chromosomes painted to at least the grandparent level.
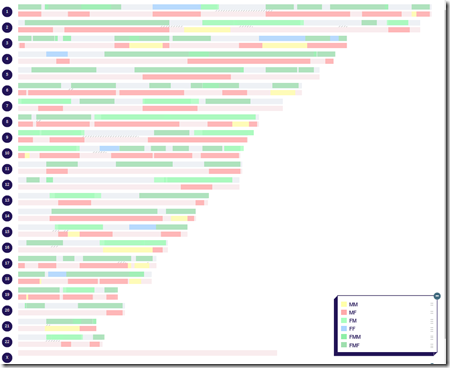
If I load this mapping into DNA Painter, it gives me:
and yahoo! I even have a bit of my one X chromosome painted to my MM side.
Confirming the Grandparent Side
This is nice. But I still have to remember that I’m not sure whether MF and MM are correct as MF and MM or if they are reversed. As it turns out, I can use clustering information that I did at Ancestry DNA to tell. Over at Ancestry, I have 14 people whose relationships I know that have tested there. And some of them are on my mother’s side.
I have previously used the Leeds Method and other clustering techniques to try to cluster my Ancestry DNA matches into grandparents. Here’s what my Leeds analysis was, with the blue, green, yellow and pink being my FF, FM, MF and MM sides.
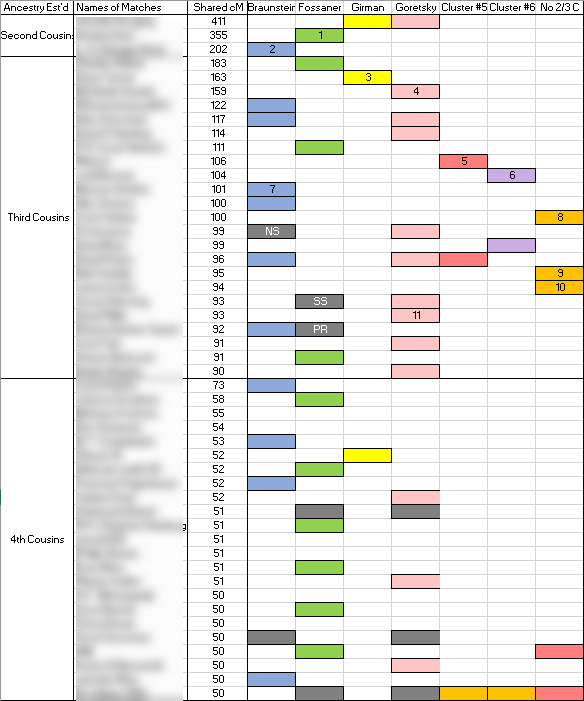
And lucky me, I did find a couple of people on GEDmatch who tested on Ancestry who were in my MM clusters from DMT. And in fact they were in my pink grouping which is my mother’s mother’s side in my Leeds method. So this does not prove, but gives me good reason to believe that I’ve got the MF and MM clusters designated correctly.
Extending Beyond Grandparents
You would think I could extend this procedure. After all, now that I have a number of people who are on the MF side, some of them should be MFM and some should be MFF.
Well I probably could, but I’d have to be careful. Because now I have to be sure the people I pick are 3rd cousins or further. Otherwise, they match me on both sides. So there is a limit here. This might be something I explore in the future. If you can understand anything of what I’ve been saying up to now, feel free to try it yourself.
Same for Paternal Grandfather
I now have people mapped to 3 of my grandparents: FM, MF and MM. I can do the same thing to get some mapped to my father’s father FF. In exactly the same way that I did it for my maternal grandparents, I can assume that my highest F match is likely FF because it would have been mapped FM if it was not. So I’ll give an MRCA of FF to Eaaaa Jaaaaaaaa Kaaaaaaaa who matches me 71 cM on 9 segments, where one of those segments triangulates with some of my B people.
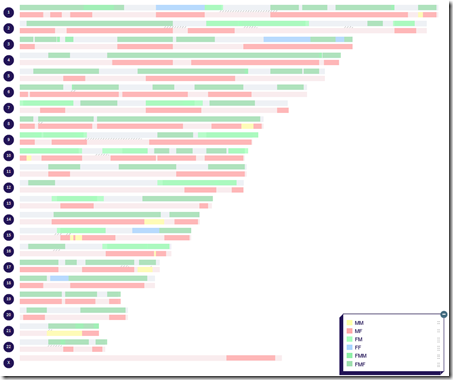
I run everything all again and I now people are clustered this way:
The FF assignments stole some people from the other Fxx groups. There are still a lot of people clustered into FMF, but that is possible. Some sides of your family may have more relatives who DNA tested, even a lot more than others do.
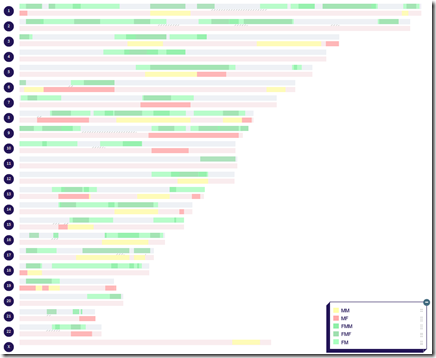
Loading my grandparent mappings into DNA Painter now gives me this:
You can see all 4 of my grandparents (FF=blue, FM=green, MF=pink, MM=yellow) and a bit more detail on my FM side.
Filling in the Entire Genome
If I had enough people who I know the MRCA for who trace back to one of my grandparents or further, and all of my triangulations with them covered the complete genome, then theoretically I’d be able to map ancestral paths completely. Endogamy does make this more difficult, because many of my DNA relatives are related on several sides. But the MRCA, because it is “most recent”, should on average pass down more segments than the other more distant ancestors do. Using a consensus approach, the MRCA segments should on average outnumber the segments of the more distant relatives and should be expected to suggest the ancestor who likely passed down the segment.
Right now in the above diagram, I have 34% of my genome mapped.
So if I go radical, and assume that DMT has got most of its cluster assignments correct, then why don’t I try copying those clusters into the MRCA column and run the whole thing again. That will allow DMT to use them in triangulations and those should cover most of my Genome. Let’s see what happens.
Doing this has increased my coverage from 34% to 60% of my genome. About 25% of the original ancestral path assignments were changed because of the new assumptions and because the “majority rules” changed.
With DMT, the more real data you include, the better the results should be. What I’m doing here is not really adding data, but telling DMT to assume that its assumption are correct. That sort of technique in simulations is called bootstrapping. It works when an algorithm is known to converge to the correct solution. I haven’t worked enough with my data to know yet whether its algorithms converge to the correct solution, so at this point, I’m still hypothesizing. The way I will be able to tell is if with different sets of data, I get very similar solutions. My matches from different testing companies likely are different enough to determine this. But I’m not sure if I have enough known MRCAs to get what would be the correct solution.
Let’s iterate a second time. I copy the ancestral paths of the 25% that were changed over to the MRCA column. This time, only 3% of the ancestral path assignments got changed. So we are converging in on a solution that at least DMT thinks makes sense.
I’ll do this one more time, copying the ancestral paths of the 3% that were changed over to the MRCA column. This time, only 1.5% changed.
Stopping here and loading into DNA Painter gives 63% coverage, remaining very similar to the previous diagram, although I’m surprised the X chromosome segment keeps popping in and out of the various diagrams I have here.
What Have I Done?
What I did above was to document and illustrate some of the experimentation I have been doing with DMT so I can see what it can do, figure out how best to use it, and hopefully map my genome in the process. Nobody has ever done this type of automated determination of ancestral paths before, not even me.
I still don’t know if the above results are mostly correct or mostly incorrect. A future blog post (Part 3 or later) will see if I can determine that.
By the way, in all this analysis, I found a few small things to fix in DMT, so feel free to download the new version 3.1.1.