In Part 1, I gave you a flavour of the mass-triangulation that I am doing, which I called EAST: Extreme Autosomal Segment Triangulation. Triangulation is a technique to determine what parts of your DNA come from what ancestors. That will help you determine how your matches are related to you.
What is new about this EAST technique is that it uses segment matches of two people (your own and a relative) rather than just one (your own).
Single Match Triangulation (SMT)
To make this clear, let me first describe the standard way people currently triangulate with FamilyTreeDNA data. I’m going with the same example I used in Part 1, using my uncle Harry as person A, and my 3rd cousin Joel as person B. They are 2nd cousin’s once removed and their most recent common ancestors are my great-great-grandparents Hirsch Focsaner and his wife Dwora.
To triangulate, I first need the segments that match between Harry and Joel. I go to Harry’s FamilyTreeDNA account, select Chromosome Bowser, and pick Joel to match to. It gives this diagram:
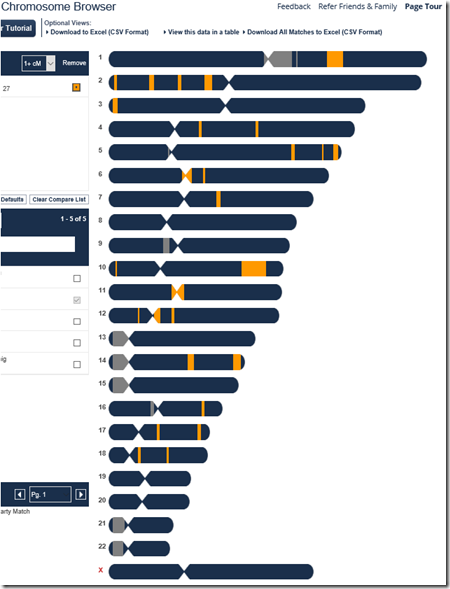
Harry and Joel match on the orange segments.
Now we need to find a relative of both Harry and Joel to be the third person of the triangulation. We go back to the FamilyTreeDNA matches page and next to Joel’s name, we click on the 4th cute little symbol below his name to “Run Common Matches”.
That brings up a second menu and we select “In Common With”. Then, if you are lucky like me, you’ll be presented with 232 pages of matches containing the 2,318 people who match to both both Harry and Joel.
Now write down the names of the top 4 and go back to the Chromosome browser and add them along with Joel. Now you’ll see:
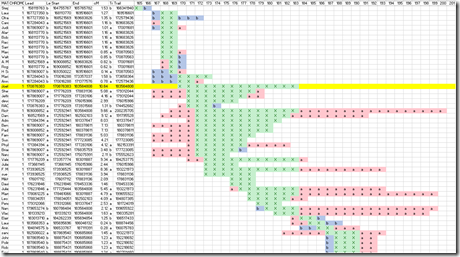
These are still Harry’s chromosomes. Joel’s matches with Harry are shown in orange. We want a third person who matches with Harry and Joel. These four people only have one instance, in chromosome 1, where one of the others matches one of Joel’s segments. You can see it in chromosome 1 as the green line that is under the orange line. Joel’s match with Harry (the orange line) is 10.64 cM. The green match is with someone named Daniel and it is 18.76 cM. So we have a triangulation. Harry matches Joel where Joel matches Daniel and Daniel match Harry.
The chromosome match setting was for a minimum 5+ cM. You could go down to the 1+ cM and you’ll find a lot more matches. But there’s a problem with this. Because of the way DNA analysis companies determine matches (that half-identical thing), there is a very good chance with small matches that they are not Identical by Descent and you don’t want that. i.e. you need them to be a true relation.
So you’ll have to stick to those 5+ cM matches to be safe.
But in the above we did find that one triangulation we can use. That third person has a segment in common with Harry and Joel. This indicates that the third person has a common ancestor with Harry and Joel. It could be Hirsch and Dwora or it could be an ancestor of Hirsch and Dwora.
So now I invite you to continue to do this for the other 2,314 common matches of Harry and Joel. You’ll tire quickly!
Doing this allows you to create Triangulation Groups, building them up person by person. Triangulation Groups put likely-related people together. The analysis of triangulation groups is complicated and has been written up elsewhere. Jim Bartlett describes it very well on his wonderful Segmentology blog, but I’m not going to get into it, because this only uses segment matches of a single person. I’m going to be doing it differently using the segment matches of two people.
Double Match Triangulation (DMT)
Just to let you know, the terms Single Match and Double Match triangulation (SMT and DMT) as well as EAST (Extreme Autosomal Segment Triangulation) are my own. I invented them so that I can talk about them. As far as I can tell, I don’t believe anyone else has extended regular triangulation this way. The closest thing I’ve seen so far is Roberta Estes’ article Just One Cousin, which used chromosome matches between three people. But I want to go from three to extreme. So let’s get into it.
The reason why SMT is referred to as “Single” match, is because only the segment matches of one person is used. Only Harry’s matches in the example above are used. Although we found the people who matched to Joel, we did not use Joel’s segment matches.
To do the Double Match Triangulation, I emailed Joel and he sent me his match list. Please see Part 1 where I describe what this file is and how to get it. I merge my uncle’s chromosome match list with Joel’s match list and I put it into Excel and add some fancy coloured mapping of the chromosomes.
Doing this for the same segment 1 region used in the above SMT example gives the following (which is the same picture I showed in Part 1):
The line in yellow is the chromosome 1 match of Harry with Joel. The green area with X’s on the yellow line is their match segment. Remember that second picture of FamilyTreeDNA’s chromosome browser from above? Look again at Chromosome 1:
The short orange line is my line in yellow. The longer green line is the is the line that is exactly 6 lines below my line in yellow belonging to Daniel. The part of that line shown in green with X’s is Daniel’s match with both Harry and Joel. The two parts on either end shown in red with a’s is Daniel’s match with Harry (but Joel doesn’t match). On other segments you can see the line in red with b’s. Those are places the third party matches to Joel but not to Harry.
What’s great about this Extreme triangulation technique is that:
- It picks out everybody who has matching segments to you AND to a selected second person. That gives all three connections needed of the triangulation triangle for everyone in a block with one of those yellow lines. This really increases the odds of the three of you being Identical By Descent (IBD). Jim Bartlett says he’s fairly confident that triangulation works down to 5 cM. Jim also says “shared segments below 5 cM are uncharted territory for triangulation.” And he was talking about Single Match Triangulation. New research about Double Match Triangulation by Michael Maglio indicates that a false positive is statistically improbable, indicating the match is IBD (or maybe IBP – identical by population, which is still IBD, but too many generations back to be of much use). So Double Match Triangulation can be used even for small segments.
- You get to see, not just all the third party segments matching to you, but also the third party matching to your second person that don’t match to you. This is additional information you don’t get from normal SMT triangulation that I’ll soon show is very useful.
- You only have 1/16 of your great-great-grandfather’s segments. But your 3rd cousin has another 1/16. With DMT, you’ve doubled the segments you can match with.
- I suspect all three connections may not be necessary. You and your cousin will only match on 1/16 of each others segments. So if you find what looks like a big Triangulation Block of known cousins, and you match to them, and your cousin matches to them, that may be good enough. I’ll have to test this, and if it works, it will make this technique another order of magnitude more powerful in classifying your matches.
- Huge time savings for analysis. One EAST is a Triangulation with every single one of your matches at once. And that’s just using one selected known relative as the second person. You can use others as well. You don’t even have to use known relatives. EAST should show you if the second person is significant within your matches..
- Lots more that I haven’t even worked out yet.
What we haven’t done yet is to use the EAST data to analyze and classify the segments of your matched people, to put them into Triangulation Groups and identify common ancestors and where everyone fits in. That will be in the next post of this series.
… One last thing:
Triple and Multiple Match Triangulation (TMT and MMT)
I want to define these now, because I see it is possible. Get the segment matches of 3 or more relatives and put them all in the same file together. Process them the same way as described in Part 1.
I don’t know if early on in the study of what EAST can do, getting into this complication is worthwhile. It will visually be hard to interpret because instead of having 3 colours (green for both match, blue for only A matches, red for only B matches), with triple match you’ll need 7 colours and Quadruple Match would need 15 colors.
It might be better to do a DMT three times (each of the three in a TMT paired three times) as each DMT would be easier to interpret than the one TMT.
But I’m getting way ahead of myself. Classifying segments will be next.
—
Follow-up June 20: Yesterday, A Triangulation Intervention was posted by Blaine Bettinger on his blog, explaining what is correct triangulation for autosomal analysis. He says:
The only way to perform true triangulation is to have segment data and a way to confirm that an overlapping segment is actually shared by two or more genetic matches.
He says the only place true triangulation tool available is the Tier 1 Triangulation tool at GEDmatch. And he says:
It is very important to note that tools like KWorks, JWorks, and ADSA at DNAGedcom, and Matching Segment Search at GEDmatch, while incredibly powerful and valuable tools, do NOT perform triangulation.
I wanted to mention this, because it’s important to understand that the tools and techniques I am developing here with EAST and DMT are all true triangulation techniques. They work with the matching segments of two people and triangulate them, not just with one or two “third people”, but with all the third people at once.
p.s. I’m building a utility program to do this EAST with DMT automatically. I expect I’ll be able to get it to classify your matches for you into true triangulation groups. It will also create comma delimited files you can import into a spreadsheet to visualize your three-way matches like I do in my Excel examples above. When the program is ready, I plan to make it available as freeware.