You’ve heard of extreme sports. Well this, I think, is the genetic genealogist’s equivalent.
In DNA or Bust, I told about getting my 93 year old uncle’s DNA analyzed. Our Endogamous Ashkenazi heritage layed 7,017 matches upon us. That was one month ago.
Since then, FamilyTreeDNA have updated their Family Finder algorithm. It was a big thing. People’s matches changed. With that change, my uncle lost 91 matches (which I had downloaded so I still have), and gained 565 matches. He still is getting a few new matches a day and is currently up to 7,611 matches. That’s quite a few.
And of those 7,611 matches, there is exactly and only 1 person in that list who I knew beforehand was a cousin. Joel is my 3rd cousin and is a second cousin once removed to my uncle, who I’ll refer to as Harry.
My common ancestor with Joel is Hirsch Focsaner and his wife Dwora Naftulovitz. They are our great-great-grandparents. Joel is listed on Harry’s Family Tree DNA list as a 2nd to 3rd cousin (so they got that right) with 134.8 cM shared and a 27.8 cM largest block.
What I want to do is: (1) Identify all the autosomal DNA segments that came from either Hirsch or Dwora, (2) Identify any people among my uncle’s 7,611 that are related to Hirsch or Dwora or to Hirsch or Dwora’s ancestors, and (3) find closer cousins in that list of matches that are descendants of Hirsch and Dwora that are related to me but not my cousin Joel, and those that are related to Joel but not to me.
And, I want to do this with all my matches together. Think it possible? Read on.
This post will talk about what I did for (1), to identify all the autosomal DNA segments that came from either Hirsch or Dwora.
Triangulation for autosomal DNA (i.e. for Family Finder tests) is a technique to use segment information of related people to determine what parts of their DNA comes from their common ancestor.
You might think you can just go to your Chromosome Browser at Family Tree DNA, and download your matching segments with your cousin, and those would be your common ancestor’s regions on your DNA.
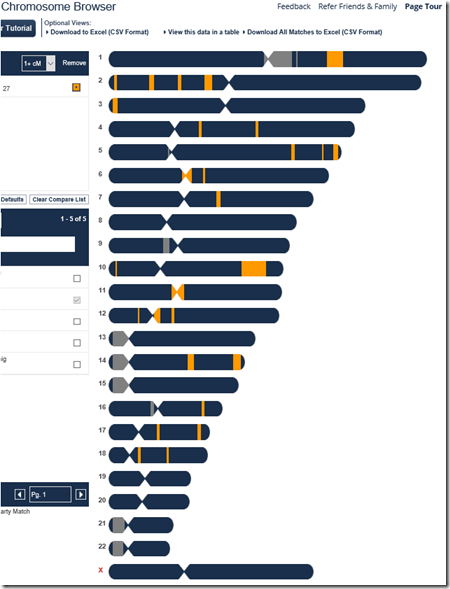
Below is my uncle Harry’s match with Joel as shown in the FamilyTreeDNA Chromosome Browser, down to the minimum 1 cM threshold. As I said above, the matches total 134.8 cM, and you can see on chromosome 10 the largest block of 27.8 cM.
That would be fine and good if it were that simple. But the way companies DNA test, they use what’s call Half-Identical Regions (HIR) which don’t guarantee Identical By Descent (IBD) matches when the matching segment size is small (e.g. under 7 cM). What is needed is to use 3 related people (a 3-way match) where person A (my uncle Harry) matches person B (my cousin Joel), and person B (Joel) matches person C (someone else who is also a descendant of our common ancestor) and person C matches person A.
You can download your Chromosome segment data into a spreadsheet from the Chromosome Browser at FamilyTreeDNA by pressing the “Download All Matches to Excel (CSV Format) button at the top right of the page (see the arrow below):
If you have a lot of matches like my uncle, it will be a big file and could take a minute before it responds, so be patient. My uncle’s file has 172,299 lines in it and is 11,820 KB (i.e. 11 MB) in size. There are on average 22.6 matches per person and the average segment match size was 3.6 cM. 3.6 cM is usually considered too small to work with because there is a good chance that those segments are not IBD. But there’s magic that happens when you triangulate with a third person, and even the small blocks become very useful.
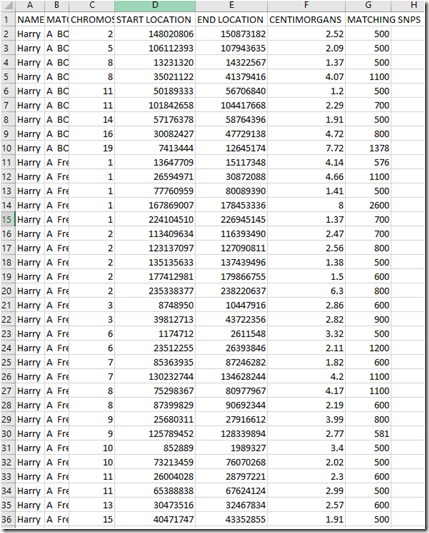
When loaded in Excel, the downloaded chromosome match file looks like this:
My uncle’s name is in the first column, followed by the person he matches to and each of the matches. The matching people are listed in alphabetical order by their full name, so all the first names are together which is somewhat awkward, but still makes it easy to find who you want.
So the above shows that my uncle matches to A BO… at 9 locations totalling 27.89 cM, and he matches A Fre… at 31 locations (only 26 shown) totalling 102.42 cM.
I can scroll down in this file and find my cousin Joel, so my uncle’s matches with him are in the file as well. This file therefore contains the A to B (Harry to Joel) matches, and the A to C (Harry to anyone else matches). But it is not good enough. I need the B to C (Joel to other people matches) to complete the triangulation.
So what I did was email my cousin and ask him if he would download all his chromosome matches and send them to me. He, of course, is as interested as me of getting something out of his DNA test, so he gladly did so.
My cousin had 73,644 lines in his file, which is less than half of what was in my uncle’s file. That might be because my uncle is one generational level higher on our tree than Joel and I are.
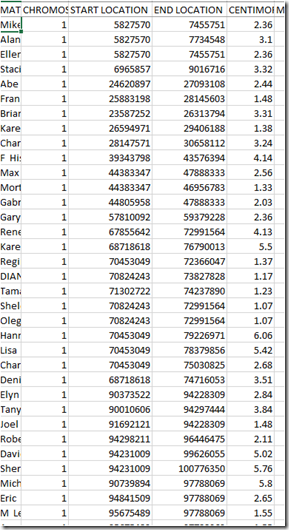
So I located my uncles chromosome matches to Joel in my uncle’s file: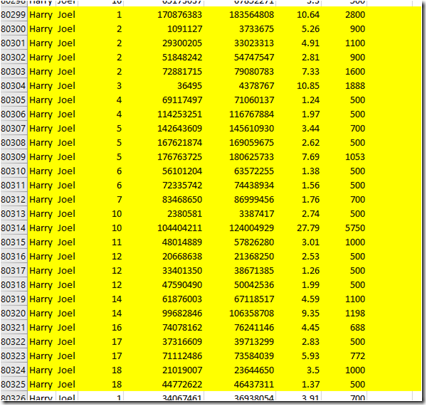
Similarly, I found Joel’s matches to my uncle in his file. It was good news. They were identical.
I combined our two files and sorted it by person and chromosome and start and end. This merged Harry and Joel’s chromosome matches together for each person. This file had almost 245,000 lines in it. Column 1 identified whether that segment was from Harry or Joel’s file.
This made it possible to find all segments for every person where both Harry and Joel matched or at least overlapped.
There were 2,041 people who had 4,582 segments that both Harry and Joel matched to. These are two-thirds triangulated, because Harry (A) matches to the third party (C) and Joel (B) matches to the third party (C).
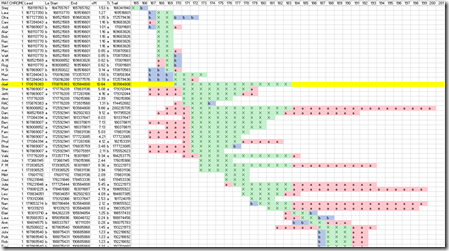
In order to do the third triangulation of Harry (A) to Joel (B), I simply added the 26 match lines (above) and left them in yellow so I could see them.
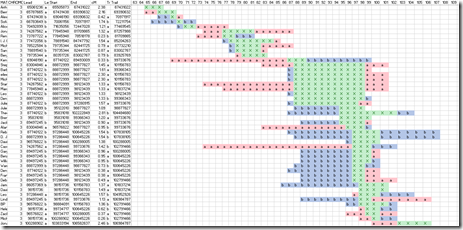
Then I made this nifty diagram out in Excel, where the column’s represent the segment location (by millions):
The green X’s are where both (A) matches (C) and (B) matches (C). The yellow line indicated where (A) and (B), my uncle and my cousin match. It guarantees that all the other matches in the same range, from 170 to 183 for all those people highlighted at the left in green, are descendants of my uncle and cousin’s common ancestors and/or their ancestors.
Once again, the green X’s again are where both my uncle matches the third party, and where my cousin matches the third party. The red a’s are where only my uncle matches the third party. The blue b’s are where only my cousin matches the third party. You can see many cases where the a’s and b’s are added on either side of the green X’s.
There are a few reasons for this that I’m still trying to sort out. Any help from genetic experts would be appreciated. But it is my understanding that:
- Every descendant only gets partial segments from their ancestors. The parts may be so different that two descendants don’t even match on the segment.
- There are actually two common ancestors, not one. My thinking is that descendants get half of Hirsch’s and half of Dwora’s, but I’m not sure if that’s the real way the DNA tests will differentiate because of the use of both genomes in the matching.
Now not all of the (A) (C) matches and (B) (C) matches have a corresponding (A) (B) match. For example:
There is no yellow bar going through here. So there is no absolute confirmation that these people are related to the common ancestor with the third part of the triangle. The segment sizes here are small (only a few cM) and my uncle and cousin just may not have been passed down the same segment parts in this area of the chromosome.
But that likely doesn’t matter. If those other 20 people have been shown to be descendants from the other triangulations with the yellow lines, then the 20 verified descendants of Hirsch and Dwora all matching together will confirm another segment from them or their ancestors. Yes, there is a chance that there is another ancestor on a different side contributing. But like triangulation, extra matches decrease greatly the odds of a false-positive. If there is another ancestor they come from, all or almost all of those 20 would have to descend from that other branch as well.
And that’s the first big take-away. When we mass-triangulate like this, we not only can get the segments that your ancestors passed down to you, but we can also reasonably accurately identify the segments they passed down to their other descendants, but not to you.
And endogamy doesn’t play into this either. It doesn’t matter if you’re a descendant of the same person in 1 way or 200 ways. You will still be getting parts of that person’s DNA.
This opens up a whole new realm for analysis.
In my next post on this, I’m going to organize all the 2,041 match people into triangulation groups and see what else we can learn about them.