One thing missing that I am adding to the upcoming version 1.2 release of Behold is an indication of relationships between people. If there is one thing not easy to determine, it is how people are related to the main person (also called the proband).
But just as importantly, once you know the relationship, there is much valuable information that can be reported that can aid in DNA research. To do so, there needs to be a concise notation for showing the relationship of one individual to another.
I am presenting my proposal for this notation here with the hope that people who are more expert at genealogical DNA research than I can comment and critique and that I can finalize a system that will be simple and will work.
Here is the basis of what I’d like to notate:
We have a person of interest in your family tree who has some sort of relationship to the proband (who is usually you, your spouse, or some relative). We want to designate the connection through male and female lines using:
- X for a female biologically related
- Y for a male biologically related
- ? for a person whose gender is unknown but biologically related
- - (that’s a hyphen), for a person who is not biologically related
Note that I am using X and Y for female and male which are the universally recognized two sex-determining chromosomes. This is better than using abbreviations for female and male (F, M) or mother, father (M, F) which is English-based and which also can lead to dumb mistakes if the incorrect interpretation is used.
So this is how I propose this notation will be written: You start with the person of interest, work up to the common ancestor (if there is one), and then back down to the proband, selecting the character to represent each person along the path.
Here’s a few examples:
My great-grandfather to me (on my mother’s side): YYXY
The first Y is my great-grandfather, the second Y is his son (my grandfather) which could be an X if this was my other great-grandfather on my mother’s side, the third character is an X for my mother and the Y at the end is me.
My great-granddaughter to me (via my daughter and grandson): XYXY
The first X is my-great-granddaughter, the Y following is her father, my grandson, then the X is my daughter, and the last Y is me.
Interestingly, the first example works down from my great-grandfather to me, but the second works up from my great-granddaughter to me. The direction doesn’t matter. The notation will always denote the path from the first person to the second.
Let’s get more complicated and include relationships that have a common ancestor:
My first cousin once removed to me: XYXYXY
Well, there’s many different ways a person can be a first cousin once removed (1c1r) to me. I’m picking just one of these possibilities with the person being the daughter of my first cousin. So this designates that my 1c1r is female, her father is my first cousin, and his mother’s father is my grandfather. And the connection is on my mother’s side.
Here, the path actually goes up to the common ancestor, and then back down to the proband. In fact, there are really two common ancestors for this line, the other one being my grandmother, and that line would be: XYXXXY with the “Y” in the fourth position being replaced by an “X”.
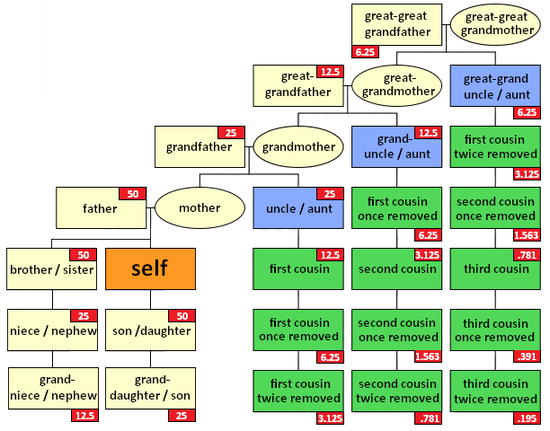
Why do we need this? Well, from the series of letters, the DNA-based relationship of the two people can be calculated. The first two examples YYXY and YXYX take 3 steps to go from the first person to the last. Each step is a sharing of 50% the autosomal DNA. That means the first and fourth people should share 50% x 50% x 50% = 12.5% of their autosomal DNA. The XYXYXY in example 3 has six steps from the first to the last. They should share 3.125% of their DNA.
That 3.125% is for the male common ancestor. If his wife/partner is also a common ancestor, then her connection adds another 3.125% and you get the total autosomal share of 6.25% for a first cousin once removed, which is what all the tables say as shown in the graphic below from DNA-explained.com:
The designation of the sex along the way is also important. All Y’s from the person of interest to your common ancestor indicate a male-line connection and you’ve found a person who could very well be a Y-DNA candidate for your common ancestor. All X’s may indicate a Mitochondrial DNA candidate for your common ancestor. Also, the exact specification of the X’s and Y’s along the way can be used to determine the percentage share of your X chromosome. Using this information, I’ll be able to get Behold to display these percentages.
The two other characters in the notation are also important. If you don’t know the sex of one person along the way, then use a question mark as their placeholder. By doing so, the length of the line is still correct and the DNA relationship percentages can still be calculated, e.g. If your 1c1r’s grandparent was Terry, but you don’t know if Terry was male or female, then you should write: XY?XXY.
The other character is a hyphen which should be used to designate a person who breaks the biological line. For example, in your genealogy you may have a cousin who was adopted. You still consider them a full cousin, and you want them documented in your family tree. But they are not of use to you in your DNA research. So the hyphen is inserted for people who break the biological connection, e.g. in this case, the parent of your cousin. Then this example would be written like this: Y-XXY.
I think this gives a lot of information in a concise easy to understand notation. I have been looking, but I have not been able to find any similar notations that have been formalized. Maybe there is something already out there that I’ve missed. If so, could you please tell me about it.
I would really appreciate your comments, ideas and suggestions and I’ll then be able to finalize this possibly new notation.
Refinement: Dec 20:
The simple notation above does not indicate the character representing the common ancestor. Often that person needs to be known, e.g. to see if there is an all-male or all-female connection to the common ancestor. I like the method suggested by Rob Hoare in the comments below to use parenthesis to surround this person. Using this, example 3 above would now be: XYX(Y)XY.
The nice thing about this extension is that, since there are always two common ancestors, a father and mother, they can both be designated together if desired, as in: XYX(YX)XY
Then in Behold, I could succinctly show the common ancestors together, e.g.:
Jane Person
Relationship: 1c1r of John Proband via Fred and Wilma Ancestor
Line: XYX(YX)XY, Shared DNA: 6.25%at, 50%X
where 6.25% is the Autosomal and 50% is the X-chromosome shared percentages between Jane Person and John Proband through this connection.
If a person is related multiple ways through different common ancestors, each relationship can easily be shown on its own line with its own DNA contributions. The DNA contributions are additive, so the total shared DNA can then be shown.
The parenthesis designation can also be used usefully to denote the direction in a direct line. The first two examples then become:
My great-grandfather to me (on my mother’s side): (Y)YXY
My great-granddaughter to me (via my daughter and grandson): XYX(Y)
Update: On May 23, 2016, I finalized and formalized the notation that I talk about in this post.

Joined: Sun, 16 Nov 2014
6 blog comments, 0 forum posts
Posted: Sat, 19 Dec 2015
Excellent idea, Louis. Lots of information in a very concise and clear format.
I think it would be useful to know there’s a most recent common ancestor in the path, and where. How many generations back to the MRCA is important (I think also there would be subtle differences in the probabilities of inheritance with three generations each side of the descent from a MRCA, compared to a run back 6 generations in a row).
A suggestion for the MRCA: surround it with angle brackets: XYX<Y>XY (shows descent directions), or (easier to parse and read?) parentheses: XYX(Y)XY.
You could show descent from two MRCA (husband and wife) like XYX<XY>XY but that’s probably overcomplex. Simpler to just have two paths like XYX(Y)XY and XYX(X)XY as (perhaps many) multiple paths will be needed anyway when there’s cousin marriages.
Joined: Sun, 16 Nov 2014
6 blog comments, 0 forum posts
Posted: Sat, 19 Dec 2015
The angle brackets in my previous comment where removed by the commenting software (it probably thinks they’re html tags), so they’re out. :-)
For example, on the “A suggestion for the MRCA” there was a Y inside greater than and less than signs before the final XY of XYXXY. Parentheses would work better.
Joined: Wed, 8 Aug 2012
10 blog comments, 0 forum posts
Posted: Sun, 20 Dec 2015
I’m glad someone is looking at this Louis, but there’s a part of the proposal that I don’t quite understand. Imagine that a DNA test shows that two people have some genetic connection. Obviously, it doesn’t imply that either one is descended from the other, but probably indicates that they have a common ancestor somewhere further back. How do you deal with that given that (a) you don’t know how far back, and (b) that common ancestor may not appear in either individual’s tree (e.g. extra-marital relationships in both cases)?
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 20 Dec 2015
I really like your ideas, Rob. I was originally thinking of maybe using a different letter, or a different color, or maybe bold text. But parenthesis is better because it can be transferred as raw text and has the advantage of being able to include both common ancestors if desired as in: XYX(XY)XY. I’ve now added a refinement at the end of the post which includes your idea.
I’ve attempted to fix the brackets and missing letters for you in your first post. Let me know if I didn’t place them correctly.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 20 Dec 2015
Tony:
Getting me to think again, aren’t you.
The purpose of this notation is to precisely define a known relationship between two people, so I wasn’t thinking of determining the relationship from the DNA.
However, the wonderful byproduct is that this notation could now make that possible. With the addition of the parenthesis refinement, every combination of letters gives exactly one set of DNA percentages. A list of these up to, say, 5 generations apart can easily be generated and then sorted by DNA percentages.
Then a person who has a DNA connection with a certain percentage Autosomal and X can then look up in the table and see what the closest matching and possible connections are.
Joined: Sun, 16 Nov 2014
6 blog comments, 0 forum posts
Posted: Sun, 20 Dec 2015
Thanks for fixing up the first comment (it did show that angle brackets was a bad choice!). Allowing more than one ancestor inside the parentheses does make the string a bit harder to parse and count: for example it’s easy to see that YY(Y)YYYY (all male descendants both sides from a male MRCA) will share Y-dna, less clear with YY(XY)YYYY.
But since in most cases (other than second marriages etc) there will be two MRCA’s, it would probably be best to allow (XY) to avoid the majority of records having two paths.
“every combination of letters gives exactly one set of DNA percentages” - these are not exact percentages, just the centre of a range of probabilities. For example, you get around 50% of dna from your mother. But it’s a random selection of her dna, and by chance some will come from her father, some from her mother. So rather than 25% from a grandparent it could be 20% and 30% (on average it will cluster around 25%).
See Blaine Bettinger’s table under “Distribution of genealogical relationships for given amounts of shared DNA” on isogg.org/wiki/Autosomal_DNA_statistics - the percentages from a grandparent are in the range 13-35%, for the previous generation 8-16% (rather than exactly 12.5%). So probabilities could be calculated, not fixed shares (but unfortunately Blaine’s raw data at thegeneticgenealogist.com/2015/05/25/the-shared-cm-project-an-update/ has a no-commercial-use restriction).
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 20 Dec 2015
Rob: I was looking into possibly giving ranges of percentages. I did look for, but was unable to find any statistics about the random nature of how DNA combines. If I had some theoretical study that estimated the combinatorial probabilities, then I might be willing to include ranges using that.
But I don’t think its right to use ranges taken from samples like the ISOGG Autosomal DNA statistics. The problem with those stats is that they are relying on the submitted trees and are assuming the relationships are 100% true. Any mistakes, as well as any additional connections through other branches will increase the apparent range. So I believe those ranges are wider than theory would state.
And then, the whole thing gets really ugly when you try to compare segments or SNPs or centiMorgans and different companies have different methodologies for those, so it might be best to keep things theoretical and simple here, and provide just the expected percentage with the knowledge that there will be a small variance around them.
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Sun, 20 Dec 2015
Actually, the bottom of: http://gcbias.org/2013/12/02/how-many-genomic-blocks-do-you-share-with-a-cousin/ gets into the sort of thing I would look for, and it describes the Poisson distribution of shared blocks in a genomic region. My stats background would allow me to do the calculations necessary.
But this is a whole other level that’s probably not worth my getting into right now.
Joined: Sat, 3 Aug 2013
7 blog comments, 0 forum posts
Posted: Mon, 21 Dec 2015
While developing fs-traversal we had a need to describe arbitrary relationships. We came up with a similar notation with letters representing each step. Though we wanted to know both the direction and gender (if possible) so we came up with:
s = son
d = daughter
c = child
m = mother
f = father
h = husband
w = wife
We didn’t choose a genderless spouse character because you won’t encounter that in FamilySearch.
We then use regex to match those strings and output a human readable string describing the relationship.
hms - husband’s mother’s son - brother-in-law
mfdd - mother’s father’s daughter’s daughter - cousin
They can be stringed together to any length:
fswmfddwm - sister-in-law’s cousin’s mother-in-law
In addition to generating human readable descriptions of arbitrary relationships, we can use a more verbose format to graph arbitrary relationships: http://genealogysystems.github.io/fs-traversal-relationship-display/
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Mon, 21 Dec 2015
Thank you for this Justin. Do you know if the system is documented and formalized anywhere, or is it an informal system you all use?.
It is a bit different in goal because I’m aiming at DNA relationship mapping for the purpose of stating just blood relationships and the expected percentage of DNA shared, so only parents and children are needed. There is no need for husband and wife because they break the chain.
And we only need to go up to the common ancestor and then down to the proband. There are no up-down-up-down-up-downs. Those would automatically cause a break in the chain and indicate no shared DNA.
Interestingly, when you are representing a simple up-down, there is a 1:1 mapping between the two systems. e.g. mfdd = X(Y)XX, but then I need to include the starting person, e.g. XX(Y)XX or YX(Y)XX because for DNA, it makes a difference if the starting person is male or female.
Joined: Sun, 16 Nov 2014
6 blog comments, 0 forum posts
Posted: Wed, 23 Dec 2015
You could combine the language-independence of your notation with the flexibility of Justin’s:
1. use upper case for going up the tree (earlier in time) or at the same level, lower case for coming down
2. indicate a marriage/partnership with = (so a wife is =X, husband =Y)
The first person is needed (that’s an addition to Justin’s), and still show biological MRCA’s in brackets (if there are any).
So you get (assuming first person is female), using Justin’s examples:
X=YXy - husband’s mother’s son - brother-in-law
XX(Y)xx - mother’s father’s daughter’s daughter - cousin
XYy=XYxx=YX - sister-in-law’s cousin’s mother-in-law
or:
y = son
x = daughter
? = child
X = mother
Y = father
=Y = husband
=X = wife
Would be better with something other than ? for unknown gender, so it can be upper/lower case, but I can’t think of one that’s language independent. Perhaps ? (unknown gender child) and ^ (unknown gender parent, rarely needed).
Very little change from your initial notation when dealing with genetic relationships (just lower case on the way down), but can handle other relationships. You’ll still know that if it contains a - or = that it’s a non-genetic relationship.
This probably now way more complex than you need!
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Thu, 24 Dec 2015
Thanks for the ideas, Rob. I was thinking about using lower case for descendants and upper case for ancestors as you suggest. Instead of “?”, I was thinking of Z and z. And that would be good for an extension of this notation.
But my specific purpose for this notation is to identify the DNA relationship. For that, up and down don’t matter. And the husband/wives don’t matter either. Only the common ancestors do. With only 4 characters for people (X, Y, ? and -) plus parenthesis for the common ancestors, all the DNA statistics can be derived and easily seen.
I’ve already started programming this into Behold. I’m going to try hard to finish it up by Boxing day.
Joined: Mon, 19 Mar 2012
7 blog comments, 9 forum posts
Posted: Tue, 5 Jan 2016
Although, again, not directly relevant to DNA, there does seem to be a bit of reinventing the wheel on how to record complex ‘transverse’ relationships here.
My favourite is Mark Forkheim’s system from over ten years ago…
http://www.forkheim.ca/family/num3.html
http://www.forkheim.ca/family/num2.html
Joined: Sun, 9 Mar 2003
291 blog comments, 245 forum posts
Posted: Tue, 5 Jan 2016
cp:
I have about 4 books on genealogy numbering in my library, including one by Richard Pence which Mark Forkham refers to. Like Mark, I’m not really enamored by any of the traditional numbering systems, especially when you are combining going up the tree and going down.
Mark’s is a good attempt, Thanks for pointing it out. If you’re only trying to figure relationships, then he shouldn’t number the children but should use say, 3 and 4 for a male and female child. Otherwise his system can’t tell you if a child is a son or a daughter.
Louis